- 1git 对象压缩及垃圾对象清理
- 2关于pom文件的相关坐标解析_pom加入的坐标依赖是啥
- 3【C语言】(7)输入输出
- 4一台电脑同时安装多个tomcat服务器教程,window同时安装tomcat7、tomcat8、tomcat9三个服务器教程_安装两个tomcat
- 5基于Java+SpringBoot+Vue前后端分离手机销售商城系统设计和实现_springboot vue手机商城
- 6c语言在面向过程有啥特点,C语言是什么
- 7Docker | Docker+Nginx部署前端项目
- 8微服务—Docker(部署)_docker部署微服务
- 9python为什么找不到csv文件_python读写csv文件的方法(还没试,先记录一下)
- 10《Git篇》01.Git看这一篇就够了_git每次提交有版本号吗
烹饪第一个U-Net进行图像分割
赞
踩
今天我们将学习如何准备计算机视觉中最重要的网络之一:U-Net。如果你没有代码和数据集也没关系,可以分别通过下面两个链接进行访问:
代码:
https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation?source=post_page-----e812e37e9cd0--------------------------------
Kaggle的MRI分割数据集:
https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation?source=post_page-----e812e37e9cd0--------------------------------
主要步骤:
1. 数据集的探索
2. 数据集和Dataloader类的创建
3. 架构的创建
4. 检查损失(DICE和二元交叉熵)
5. 结果
数据集的探索
我们得到了一组(255 x 255)的MRI扫描的2D图像,以及它们相应的必须将每个像素分类为0(健康)或1(肿瘤)。
这里有一些例子:
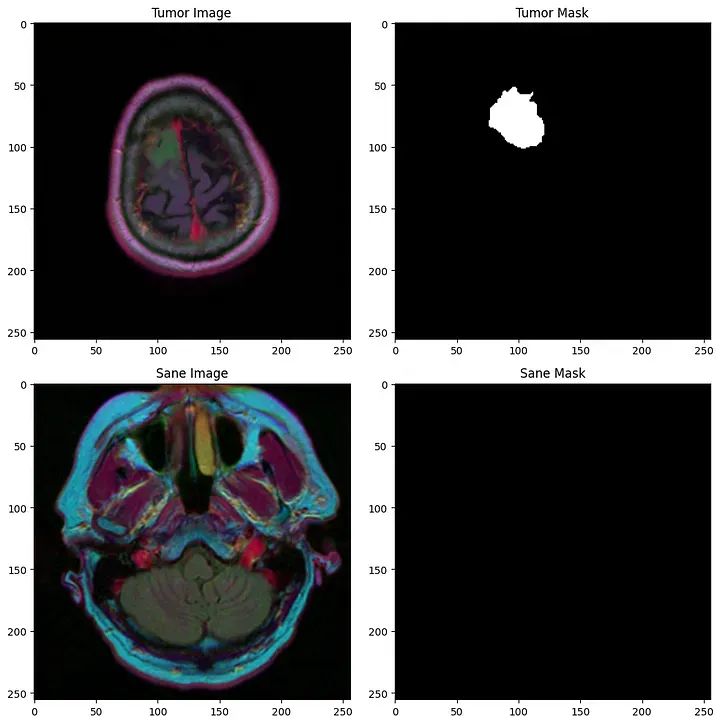
第一行:肿瘤,第二行:健康主题
数据集和Dataloader类
这是涉及神经网络的每个项目中都会找到的一步。
数据集类
- import torch
- import torch.nn as nn
- import torch.nn.functional as F
- from torch.utils.data import Dataset, DataLoader
-
-
- class BrainMriDataset(Dataset):
- def __init__(self, df, transforms):
- # df contains the paths to all files
- self.df = df
- # transforms is the set of data augmentation operations we use
- self.transforms = transforms
-
- def __len__(self):
- return len(self.df)
-
- def __getitem__(self, idx):
- image = cv2.imread(self.df.iloc[idx, 1])
- mask = cv2.imread(self.df.iloc[idx, 2], 0)
-
- augmented = self.transforms(image=image,
- mask=mask)
-
- image = augmented['image'] # Dimension (3, 255, 255)
- mask = augmented['mask'] # Dimension (255, 255)
-
-
- # We notice that the image has one more dimension (3 color channels), so we have to one one "artificial" dimension to the mask to match it
- mask = np.expand_dims(mask, axis=0) # Dimension (1, 255, 255)
-
- return image, mask

数据加载器
既然我们已经创建了Dataset类来重新整形张量,我们首先需要定义训练集(用于训练模型),验证集(用于监控训练并避免过拟合),以及测试集,最终评估模型在未见数据上的性能。
U-Net架构
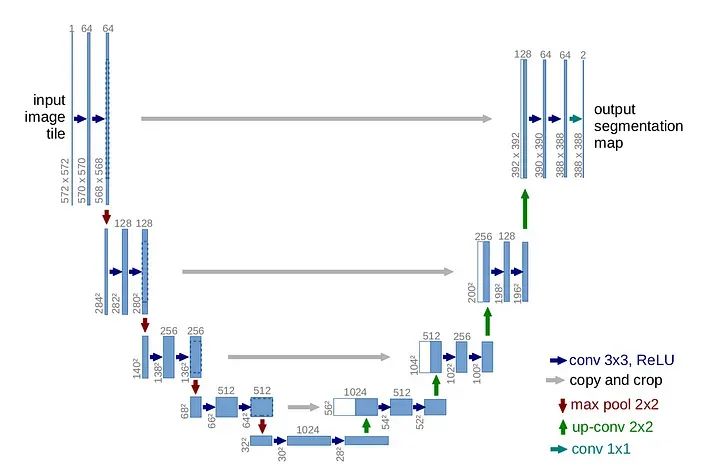
U-Net架构是用于图像分割任务的强大模型,是卷积神经网络(CNN)的一种类型,其名称来自其U形状的结构。U-Net最初由Olaf Ronneberger等人在2015年的论文中首次开发,标题为“U-Net:用于生物医学图像分割的卷积网络”。
其结构涉及编码(降采样)路径和解码(上采样)路径。U-Net至今仍然是一个非常成功的模型,其成功来自两个主要因素:
1. 对称结构(U形状)
2. 前向连接(图片上的灰色箭头)
前向连接的主要思想是,随着我们在层中越来越深入,我们会失去有关原始图像的一些信息。然而,我们的任务是对图像进行分割,我们需要精确的图像来对每个像素进行分类。这就是为什么我们在对称解码器层的每一层中重新注入图像的原因。以下是通过Pytorch实现的代码:
- train_dataset = BrainMriDataset(train_df, transforms=transforms)
- train_dataloader = DataLoader(train_dataset, batch_size=32, shuffle=True)
-
-
- val_dataset = BrainMriDataset(val_df, transforms=transforms)
- val_dataloader = DataLoader(val_dataset, batch_size=32, shuffle=False)
-
-
- test_dataset = BrainMriDataset(test_df, transforms=transforms)
- test_dataloader = DataLoader(test_dataset, batch_size=32, shuffle=False)
-
-
-
-
- class UNet(nn.Module):
-
-
- def __init__(self):
- super().__init__()
-
-
- # Define convolutional layers
- # These are used in the "down" path of the U-Net,
- # where the image is successively downsampled
- self.conv_down1 = double_conv(3, 64)
- self.conv_down2 = double_conv(64, 128)
- self.conv_down3 = double_conv(128, 256)
- self.conv_down4 = double_conv(256, 512)
-
-
- # Define max pooling layer for downsampling
- self.maxpool = nn.MaxPool2d(2)
-
-
- # Define upsampling layer
- self.upsample = nn.Upsample(scale_factor=2, mode='bilinear', align_corners=True)
-
-
- # Define convolutional layers
- # These are used in the "up" path of the U-Net,
- # where the image is successively upsampled
- self.conv_up3 = double_conv(256 + 512, 256)
- self.conv_up2 = double_conv(128 + 256, 128)
- self.conv_up1 = double_conv(128 + 64, 64)
-
-
- # Define final convolution to output correct number of classes
- # 1 because there are only two classes (tumor or not tumor)
- self.last_conv = nn.Conv2d(64, 1, kernel_size=1)
-
-
- def forward(self, x):
- # Forward pass through the network
-
-
- # Down path
- conv1 = self.conv_down1(x)
- x = self.maxpool(conv1)
- conv2 = self.conv_down2(x)
- x = self.maxpool(conv2)
- conv3 = self.conv_down3(x)
- x = self.maxpool(conv3)
- x = self.conv_down4(x)
-
-
- # Up path
- x = self.upsample(x)
- x = torch.cat([x, conv3], dim=1)
- x = self.conv_up3(x)
- x = self.upsample(x)
- x = torch.cat([x, conv2], dim=1)
- x = self.conv_up2(x)
- x = self.upsample(x)
- x = torch.cat([x, conv1], dim=1)
- x = self.conv_up1(x)
-
-
- # Final output
- out = self.last_conv(x)
- out = torch.sigmoid(out)
-
-
- return out

损失和评估标准
与每个神经网络一样,都有一个目标函数,一种损失,我们通过梯度下降最小化它。我们还引入了评估标准,它帮助我们训练模型(如果它在连续的3个时期中没有改善,那么我们停止训练,因为模型正在过拟合)。从这一段中有两个主要要点:
1. 损失函数是两个损失函数的组合(DICE损失,二元交叉熵)
2. 评估函数是DICE分数,不要与DICE损失混淆
DICE损失:
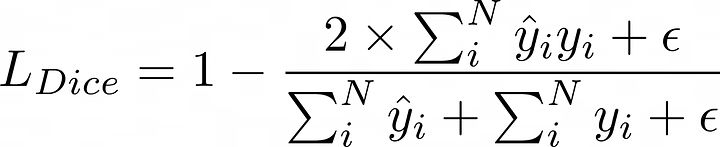
DICE损失
备注:我们添加了一个平滑参数(epsilon)以避免除以零。
二元交叉熵损失:
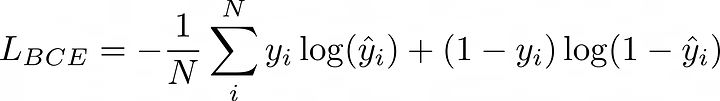
BCE
于是,我们的总损失是:
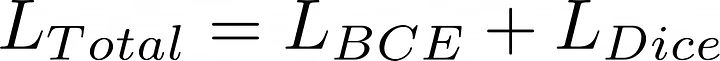
让我们一起实现它:
评估标准(Dice系数):
我们使用的评估函数是DICE分数。它在0到1之间,1是最好的。
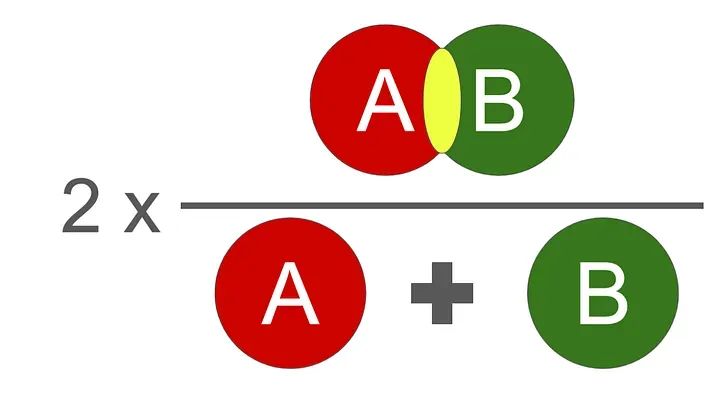
Dice分数的图示
其数学实现如下:
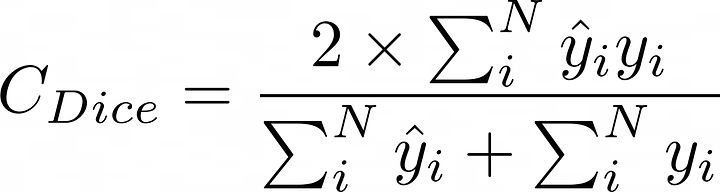
- def dice_coef_metric(inputs, target):
- intersection = 2.0 * (target * inputs).sum()
- union = target.sum() + inputs.sum()
- if target.sum() == 0 and inputs.sum() == 0:
- return 1.0
-
-
- return intersection / union
训练循环
结果
让我们在一个带有肿瘤的主题上评估我们的模型:
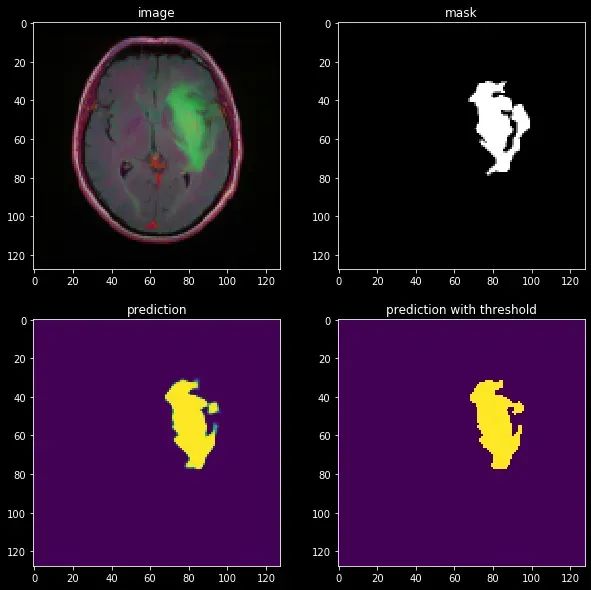
结果看起来相当不错!我们可以看到模型明显学到了关于图像结构的一些有用信息。然而,它可能可以更好地细化分割,这可以通过我们将很快讨论的更先进的技术来实现。U-Net至今仍然广泛使用,但有一个著名的模型达到了最先进的性能,称为nn-UNet。
· END ·
HAPPY LIFE

本文仅供学习交流使用,如有侵权请联系作者删除


