- 1我们好好聊聊华为Harmony OS,鸿蒙对于智能家居来说意味着什么?_harmonyos价值主张
- 2深度学习~循环神经网络RNN, LSTM_rnn lstm
- 3Spring通过IOC创建Bean的几种方式_springioc对于javabean的创建有哪几种方式
- 4人工智能AI需要什么配置的服务器开发搭建架设_人工智能开发本地要配gpu吗
- 5Netty之Marshalling编解码_netty 无法解析符号 'io
- 6程序员常用单词词汇汇总
- 7Android Gradle Composing builds 管理三方依赖_build.gradle.kts
- 8Dynamic Graph Neural Networks for Sequential Recommendation
- 9中文编程语言_纯中文编程语言
- 10开发一款Android App,从零开始详细讲解_安卓 app开发
AGNES_agnes算法
赞
踩
AGNES(AGglomerative NESting 的简写)是一种采用自底向上聚合策略的层次聚类算法。
【工作过程】:
- 先将数据集中的每个样本看作一个初始聚类簇;
- 然后在算法运行的每一步中找出距离最近的两个聚类簇进行合并;
- 步骤(2)不断重复,直至达到预设的聚类簇的个数。
【关键】:如何计算聚类簇之间的距离。
实际上,每个簇是一个样本集合,因此,只需采用关于集合的某种距离即可。
最小距离:
d
m
i
n
(
C
i
,
C
j
)
=
m
i
n
x
∈
C
i
,
z
∈
C
j
d
i
s
t
(
x
,
z
)
,
最大距离:
d
m
a
x
(
C
i
,
C
j
)
=
m
a
x
x
∈
C
i
,
z
∈
C
j
d
i
s
t
(
x
,
z
)
,
平均距离:
d
a
v
g
(
C
i
,
C
j
)
=
1
∣
C
i
∣
∣
C
j
∣
∑
x
∈
C
i
∑
z
∈
C
j
d
i
s
t
(
x
,
z
)
.
\text{最小距离:}d_{min}(C_i, C_j) = min_{x\in C_i,z\in C_j}dist(x,z), \\ \text{最大距离:}d_{max}(C_i, C_j) = max_{x\in C_i,z\in C_j}dist(x,z), \\ \text{平均距离:}d_{avg}(C_i, C_j) = \frac{1}{|C_i||C_j|}\sum_{x\in C_i}\sum_{z\in C_j}dist(x,z).
最小距离:dmin(Ci,Cj)=minx∈Ci,z∈Cjdist(x,z),最大距离:dmax(Ci,Cj)=maxx∈Ci,z∈Cjdist(x,z),平均距离:davg(Ci,Cj)=∣Ci∣∣Cj∣1x∈Ci∑z∈Cj∑dist(x,z).
显然,最小距离由两个簇的最近样本决定,最大距离由两个簇的最远样本决定,而平均距离则由两个簇的所有样本共同决定。
当聚类簇距离为
- d m i n d_{min} dmin:AGNES 算法被称为“单链接”(single-linkage);
- d m a x d_{max} dmax:AGNES 算法被称为“全链接”(complete-linkage);
- d a v g d_{avg} davg:AGNES 算法被称为“均链接”(average-linkage)。
此外,豪斯多夫距离(Hausdorff distance)也可用于集合间的距离计算。关于豪斯多夫距离的介绍可参考这篇博文豪斯多夫距离
【算法描述】:
- 输入:样本集 D = { x 1 , x 2 , ⋯   , x n } D = \{x_1, x_2, \cdots, x_n\} D={x1,x2,⋯,xn};聚类簇距离度量函数 dist;聚类簇数 k。
- 输出:簇划分 C = { C 1 , C 2 , ⋯   , C k } C = \{C_1, C_2, \cdots, C_k\} C={C1,C2,⋯,Ck}。
- 过程:
- 为每个样本创建一个簇;
- 计算距离矩阵;
- 开始合并簇过程,初始化聚类簇个数 q = n:
- 每次从距离矩阵中找出距离最近的两个聚类簇 C i C_i Ci 和 C j C_j Cj,i < j;
- 合并这两个簇(优先合并到下标较小的簇 C i C_i Ci) C i = C i ⋃ C j C_i = C_i \bigcup C_j Ci=Ci⋃Cj;
- 将聚类簇重新编号(合并到下标较小的簇可以减少重编号的次数);
- 删除距离矩阵的第 j 行与第 j 列;
- 计算合并后的簇 C i C_i Ci 与剩余其他簇之间的距离,并更新距离矩阵。
- q = q - 1。
- 直到 q == k 时,退出循环。
- 返回簇划分。
案例说明
| id | density | sugar content | id | density | sugar content | id | density | sugar content |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.697 | 0.460 | 11 | 0.245 | 0.057 | 21 | 0.748 | 0.232 |
| 2 | 0.774 | 0.376 | 12 | 0.343 | 0.099 | 22 | 0.714 | 0.346 |
| 3 | 0.634 | 0.264 | 13 | 0.639 | 0.161 | 23 | 0.483 | 0.312 |
| 4 | 0.608 | 0.318 | 14 | 0.657 | 0.198 | 24 | 0.478 | 0.437 |
| 5 | 0.556 | 0.215 | 15 | 0.360 | 0.370 | 25 | 0.525 | 0.369 |
| 6 | 0.403 | 0.237 | 16 | 0.593 | 0.042 | 26 | 0.751 | 0.489 |
| 7 | 0.481 | 0.149 | 17 | 0.719 | 0.103 | 27 | 0.532 | 0.472 |
| 8 | 0.437 | 0.211 | 18 | 0.359 | 0.188 | 28 | 0.473 | 0.376 |
| 9 | 0.666 | 0.091 | 19 | 0.339 | 0.241 | 29 | 0.725 | 0.445 |
| 10 | 0.243 | 0.267 | 20 | 0.282 | 0.257 | 30 | 0.446 | 0.459 |
以上述数据集(《机器学习》西瓜集 4.0)为例,令 AGNES 算法一直执行到所有样本出现在同一个簇中,即 k = 1,即可得到下图所示的“树状图”(dendrogram),其中每层链接一组聚类簇。
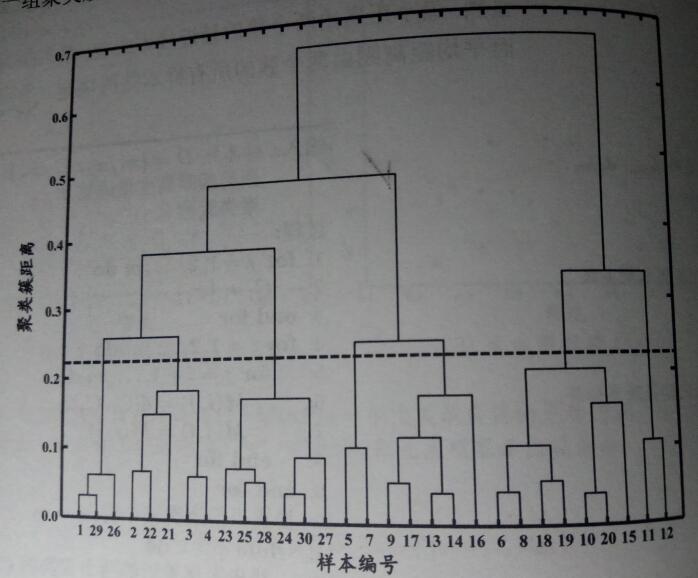
观测树状图的合并结果,可以发现该合并结果是 AGNES 算法以“最大距离”作为距离度量标准进行合并。此外,在树状图的特定层次上进行分割,则可得到相应的簇划分结果。例如,以图中所示虚线分割树状图,将得到包含 7 个聚类簇的结果。
- C1 = {x1, x26, x29};
- C2 = {x2, x3, x4, x21, x22};
- C3 = {x23, x24, x25, x27, x28, x30};
- C4 = {x5, x7};
- C5 = {x9, x13, x14, x16, 17};
- C6 = {x6, x8, x10, x5, x18, x19, x20};
- C7 = {x11, x12}。
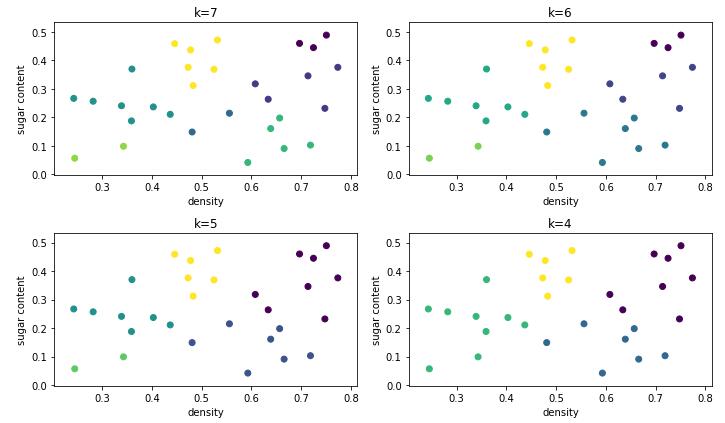
将分割层逐步提升,则可得到聚类簇逐渐减小的聚类结果。下图显示 AGNES 算法产生 7 至 4 个聚类簇的划分结果。
代码实现
首先实现距离计算函数。
def get_dist(XA, XB, type='min'):
if len(XA.shape) == 1:
XA = np.array([XA])
if len(XB.shape) == 1:
XB = np.array([XB])
dist = 0
if type == 'min':
dist = cdist(XA, XB, 'euclidean').min()
elif type == 'max':
dist = cdist(XA, XB, 'euclidean').max()
else:
dist = cdist(XA, XB, 'euclidean').sum() / XA.shape[0] / XB.shape[0]
return dist
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
然后编写 AGNES 函数的实体:
- 函数定义
def AGNES(dataset, k, dist_method='avg'):
- 1
- 初始化所需变量
# 获取样本集长度
length = dataset.shape[0]
# 初始化聚类簇和距离矩阵
clusters, dist_matrix = [], np.mat(np.zeros((length, length)))
- 1
- 2
- 3
- 4
- 为每个样本分配一个聚类簇
for data in dataset:
clusters.append(data)
- 1
- 2
- 计算距离矩阵
for i in range(length):
for j in range(length):
if i == j:
dist = np.inf
else:
dist = get_dist(clusters[i], clusters[j], dist_method)
dist_matrix[i, j] = dist
dist_matrix[j, i] = dist
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 设置当前聚类簇的个数,并开始合并过程
cluster_count = length
while cluster_count > k:
- 1
- 2
- 找出距离最近的两个聚类簇
first, second = np.where(dist_matrix == dist_matrix.min())[0]
- 1
- 合并这两个聚类簇
clusters[first] = np.vstack((cluters[first], clusters[second]))
- 1
- 将聚类簇重新编号
for i in range(second + 1, cluster_count):
clusters[i - 1] = clusters[i]
clusters.pop()
- 1
- 2
- 3
- 删除距离矩阵的第 second 行与列
dist_matrix = np.delete(dist_matrix, second, axis=0)
dist_matrix = np.delete(dist_matrix, second, axis=1)
- 1
- 2
- 重新计算距离矩阵第 first 簇与其他簇之间距离
for i in range(cluster_count - 1):
if first == i:
dist = np.inf
else:
dist = get_dist(clusters[first], clusters[i], dist_method)
dist_matrix[first, i] = dist
dist_matrix[i, first] = dist
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- q = q - 1
cluster_count -= 1
- 1
- 返回簇划分
return clusters
- 1
【完整代码】:传送门
def AGNES(dataset, k, dist_method='avg'): length = dataset.shape[0] clusters = [] dist_matrix = np.mat(np.zeros((length, length))) for data in dataset: clusters.append(data) for i in range(length): for j in range(length): if i == j: dist = np.inf else: dist = get_dist(clusters[i], clusters[j], dist_method) dist_matrix[i, j] = dist dist_matrix[j, i] = dist # 设置当前聚类簇的个数 cluster_count = length while cluster_count > k: # 找出距离最近的两个聚类簇 first, second = np.where(dist_matrix == dist_matrix.min())[0] # 合并这两个聚类簇 clusters[first] = np.vstack((clusters[first], clusters[second])) # 重新编号聚类簇 for i in range(second + 1, cluster_count): clusters[i - 1] = clusters[i] clusters.pop() # 删除距离矩阵的第 second 行与列 dist_matrix = np.delete(dist_matrix, second, axis=0) dist_matrix = np.delete(dist_matrix, second, axis=1) # 重新计算距离矩阵第 first 簇与其他簇之间距离 for i in range(cluster_count - 1): if first == i: dist = np.inf else: dist = get_dist(clusters[first], clusters[i], dist_method) dist_matrix[first, i] = dist dist_matrix[i, first] = dist cluster_count -= 1 return clusters
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
参考
- 《机器学习》周志华
- 豪斯多夫距离:https://blog.csdn.net/swallowwd/article/details/81538726



