热门标签
热门文章
- 1utf8和utf8mb4的区别_c utf8mb4
- 2【uni-app】封装websokcet以及使用方法
- 3uniapp开发的h5如何跳转回uniapp开发的app或小程序以及getLocation授权_uniapp h5返回app
- 4关于Nand Flash行地址和列地址的计算_0xffffff在nand寄存器如何排布row和column
- 5华为gt3智能跑步计划体验:心率监测、gps轨迹、训练计划_华为运动健康马拉松训练计划
- 6自动化生成代码_syetem verilog+如何生成自动化的脚本
- 7苹果手机fiddler抓包时出现了tunnel to 443 解决方案,亲测有效_苹果抓包证书被检测出来了
- 8【实习】大三暑假实习总结:工作记录、个人日记、感悟总结_allowinput=true标签的作用
- 9使用OPENCV简单实现具有肤质保留功能的磨皮增白算法_ffmpeg opencv 磨皮美白
- 10Cocoscreator2.4.7打包的IOS原生项目在IOS17+xcode15下启动崩溃_xcode15 cocoscreator
当前位置: article > 正文
Python 利用MNE实现自定义矩阵大脑拓扑图的绘制_python利用mne画脑电地形图
作者:花生_TL007 | 2024-03-11 18:02:35
赞
踩
python利用mne画脑电地形图
目的:利用MNE实现自定义矩阵大脑拓扑图的绘制
文章目录
0、加载python库
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import mne
%matplotlib qt
- 1
- 2
- 3
- 4
- 5
- 6
1、获取可用的电极布局系统
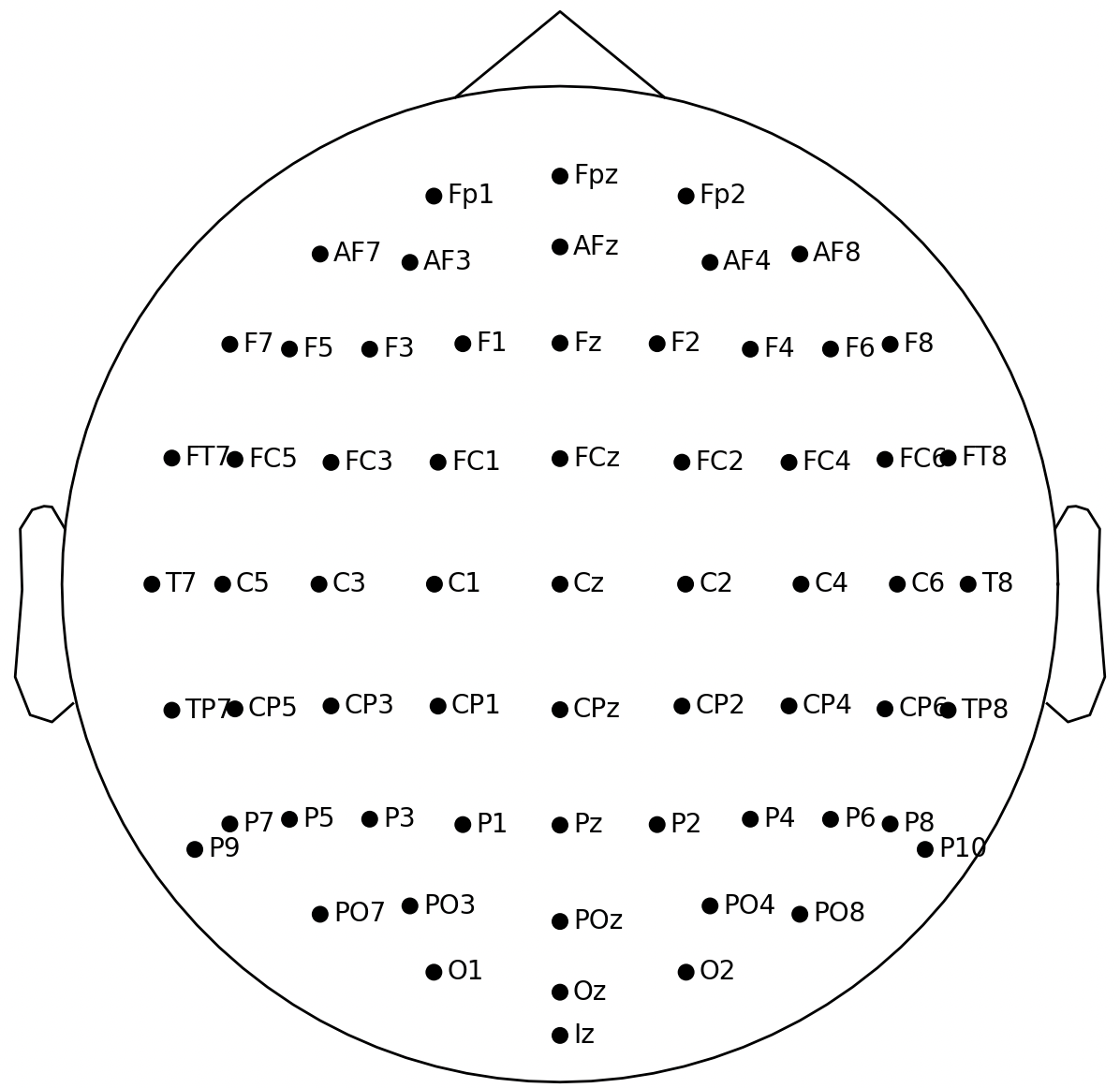
在使用MNE进行EEG信号的可视化操作时,往往需要导入对应电极的位置信息,MNE中有内置的常见电极布局系统,通过调用下面指令进行导入:
mne.channels.get_builtin_montages() # output: ['standard_1005', 'standard_1020', 'standard_alphabetic', 'standard_postfixed', 'standard_prefixed', 'standard_primed', 'biosemi16', 'biosemi32', 'biosemi64', 'biosemi128', 'biosemi160', 'biosemi256', 'easycap-M1', 'easycap-M10', 'EGI_256', 'GSN-HydroCel-32', 'GSN-HydroCel-64_1.0', 'GSN-HydroCel-65_1.0', 'GSN-HydroCel-128', 'GSN-HydroCel-129', 'GSN-HydroCel-256', 'GSN-HydroCel-257', 'mgh60', 'mgh70', 'artinis-octamon', 'artinis-brite23', 'brainproducts-RNP-BA-128']
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
从上面的输出可以看出,MNE中共有27个可用的电极布局系统,具体选择哪一个要看你采集数据时使用的脑电帽电极布局系统是哪个。
2、利用MNE自带的电极布局系统对矩阵进行通道定位
2.1 加载脑地形图位置坐标并可视化
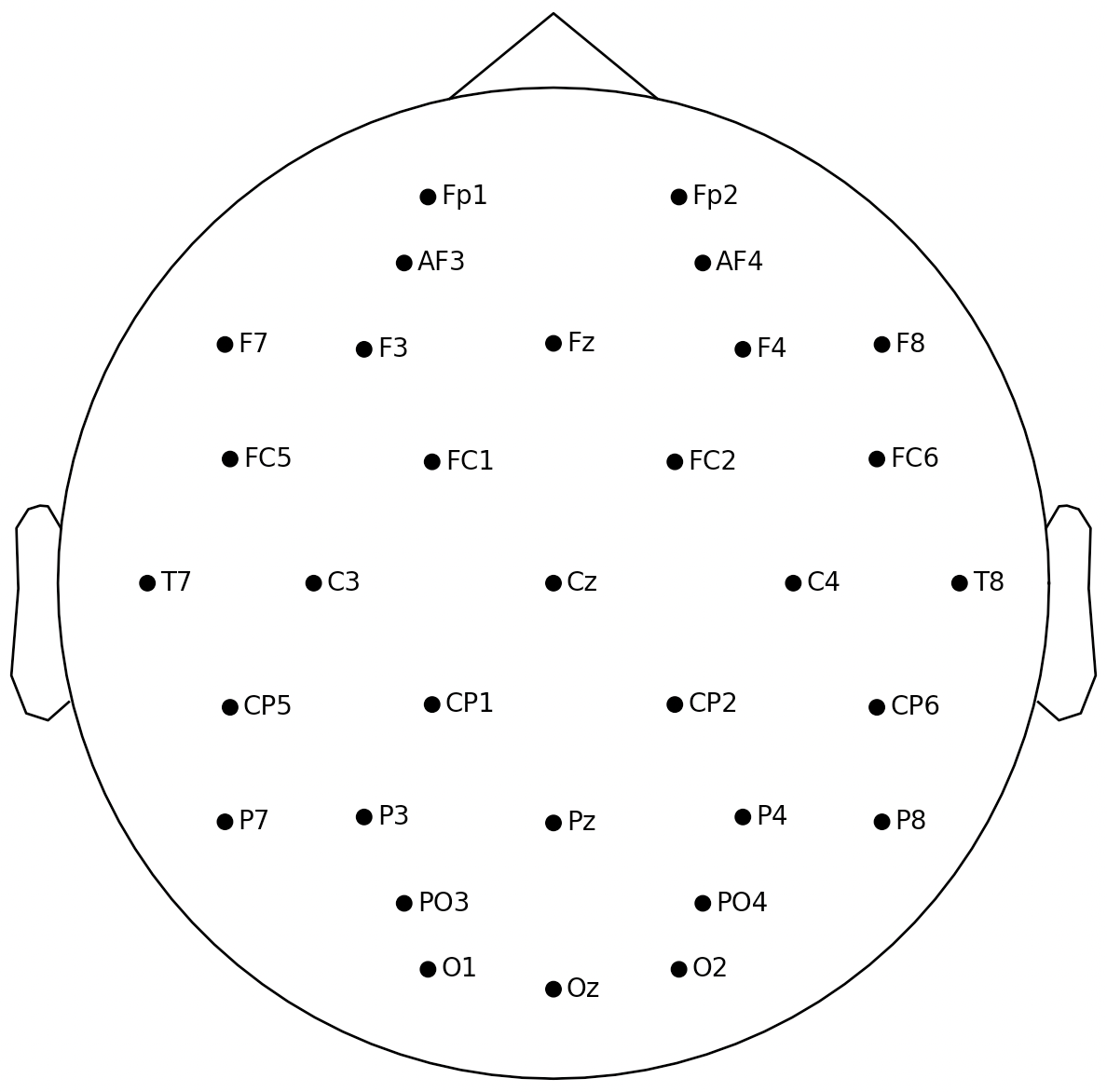
int32_montage = mne.channels.make_standard_montage('biosemi32')
int32_montage.plot()
plt.show()
- 1
- 2
- 3
2.2 构建自定义32导联的字典,导联名称与导联权重一一对应
weight = {'Fp1': 0.31, 'AF3': -0.26, 'F7': 1.22, 'F3': 0.99, 'Fz': 0.71, 'FC5': 0.55, 'FC1': -1.19, 'T7': 0.61, 'C3': -0.80, 'Cz': 2.36, 'CP5': -0.74, 'CP1': 0.72, 'P7': 0.93, 'P3': 0.38, 'Pz': 1.07, 'PO3': -1.46, 'O1': -0.12, 'Oz': 1.074, 'Fp2': 1.04, 'AF4': -0.065, 'F4': -0.52, 'F8': 0.37, 'FC2': 1.30, 'FC6': 0.94, 'C4': -1.11, 'T8': -0.16, 'CP2': 1.82, 'CP6': 0.41, 'P4': 0.46, 'P8': 0.99, 'PO4':0.15, 'O2':0.23}
- 1
2.3 根据脑地形图导联顺序重构自定义矩阵(*注意)
# 查看脑地形图矩阵导联位置
sensor_data_32 = int32_montage.get_positions()['ch_pos']
sensor_dataframe_32 = pd.DataFrame(sensor_data_32).T
chLa_index = sensor_dataframe_32.index.values
print(chLa_index)
# 重构自定义矩阵顺序
reWeight = []
for key in chLa_index:
val = weight[key]
reWeight.append(val)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
2.4 TopoMap可视化
2.4.1 创建info
info = mne.create_info(
ch_names = chLa_index,
ch_types = ['eeg']*32, # 通道个数
sfreq = 1000) # 采样频率
info.set_montage(int32_montage)
- 1
- 2
- 3
- 4
- 5
2.4.2 可视化
im, cn = mne.viz.plot_topomap(reWeight,
info,
names = chLa_index.tolist(),
# vlim=(-2, 2)
)
plt.colorbar(im)
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
3、自定义电极布局文件对矩阵进行通道定位
假设我有一个64导联的帽子,根据实验需求,实验过程中我只选择中央区和顶叶区域的电极。
那么我们该如何根据自己选择的导联制定通道定位模版呢?
这里有两种方法:
第一种:自定义新的电极布局文件,文件只包含实验选择的导联。这个方法的目的是教会你如何制作自己的电极布局文件,实际绘制时建议选择第二种方法。
第二种:使用原来的电极布局文件,先对矩阵进行归一化,后将未被选择的导联值置为0
4、第一种方法
4.1 下载标准的64通道电极布局系统坐标
# 读取MNE中biosemi64电极位置信息
biosemi_montage = mne.channels.make_standard_montage('biosemi64')
sensor_data_64 = biosemi_montage.get_positions()['ch_pos']
sensor_dataframe_64 = pd.DataFrame(sensor_data_64).T
sensor_dataframe_64.to_excel('sensor_dataframe_64.xlsx')
- 1
- 2
- 3
- 4
- 5
4.2 根据下载文件,选择实验选取的电极坐标,保存为excel格式
# 模仿下载的电极布局excel,自定义电极布局文件
- 1
4.3 加载自定义的excel文件,制作自己的montage
myStardart = pd.read_excel('mySensor_dataframe.xlsx', index_col=0) # 读取自己的电极定位文件
ch_names = np.array(myStardart.index) # 电极名称
position = np.array(myStardart) # 电极坐标位置
sensorPosition = dict(zip(ch_names, position)) # 制定为字典的形式
myMontage = mne.channels.make_dig_montage(ch_pos=sensorPosition)
myMontage.plot()
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
4.4 自定义导联权重字典
myWeight = {'C1': 0.31, 'C3': 1.22, 'P6': 0.71, 'P4': 0.55,'P2': -1.19, 'CP5': 0.61,
'CP3': -0.80, 'CP1': 2.36, 'P1': -0.74, 'P3': 0.72,'P5': 0.93, 'Pz': -1.46,
'CPz': -0.12, 'Cz': 1.074,'C5':2.34,'C2': 1.04, 'C4': -0.065, 'C6': -0.52,
'CP2': 0.37, 'CP4': 1.30, 'CP6': 0.94}
- 1
- 2
- 3
- 4
4.5 根据脑地形图导联顺序重构自定义矩阵(*注意)
# 查看脑地形图矩阵导联位置
my_chLa_index = ch_names.tolist()
print('脑地形图矩阵导联顺序:',my_chLa_index)
# 重构自定义矩阵顺序
reMyWeight = []
for key in my_chLa_index:
val = myWeight[key]
reMyWeight.append(val)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
4.6 TopoMap可视化
4.6.1 创建info
myinfo = mne.create_info(
ch_names = my_chLa_index,
ch_types = ['eeg']*21, # 通道个数
sfreq = 1000) # 采样频率
myinfo.set_montage(myMontage)
- 1
- 2
- 3
- 4
- 5
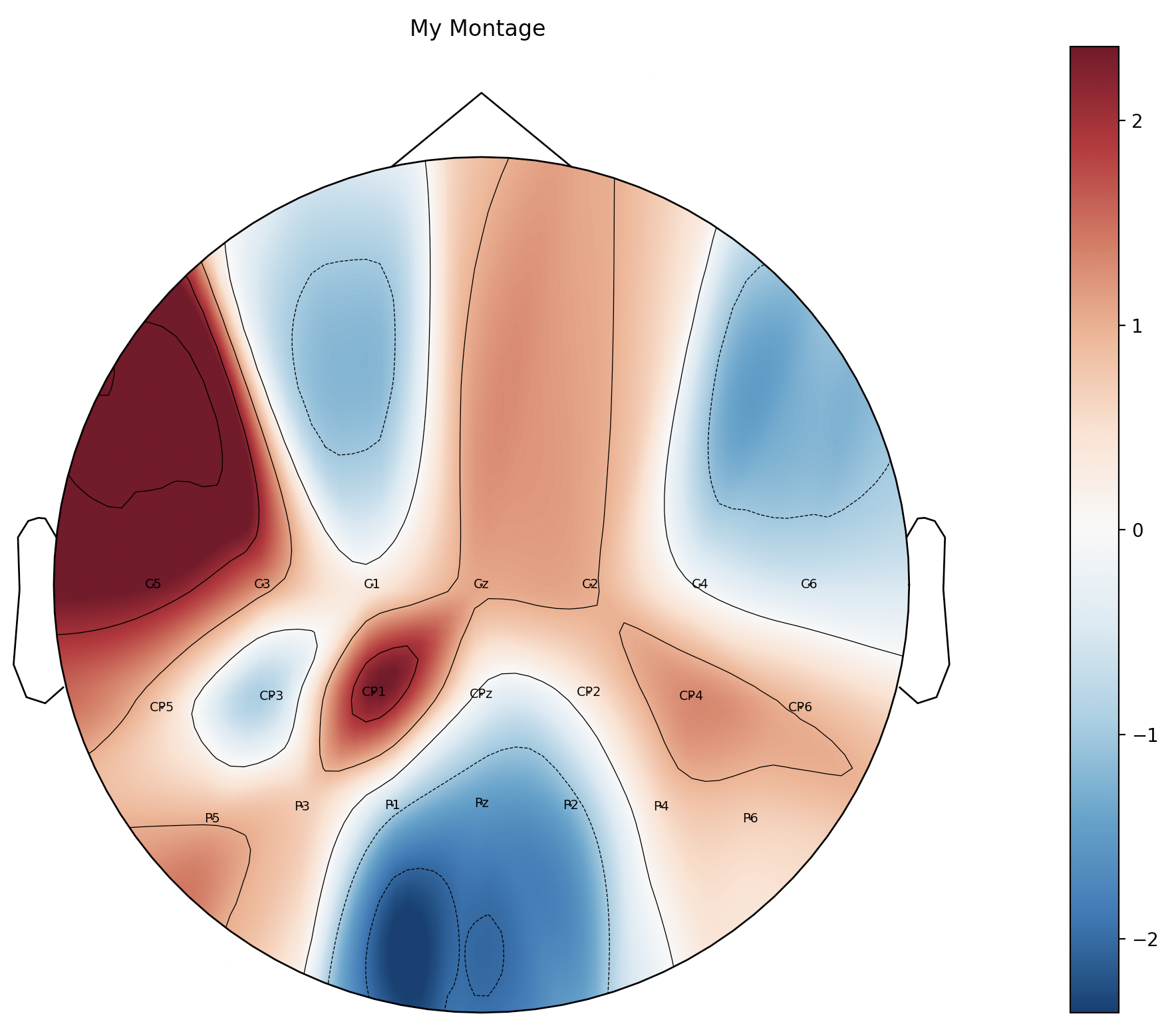
4.6.2 可视化
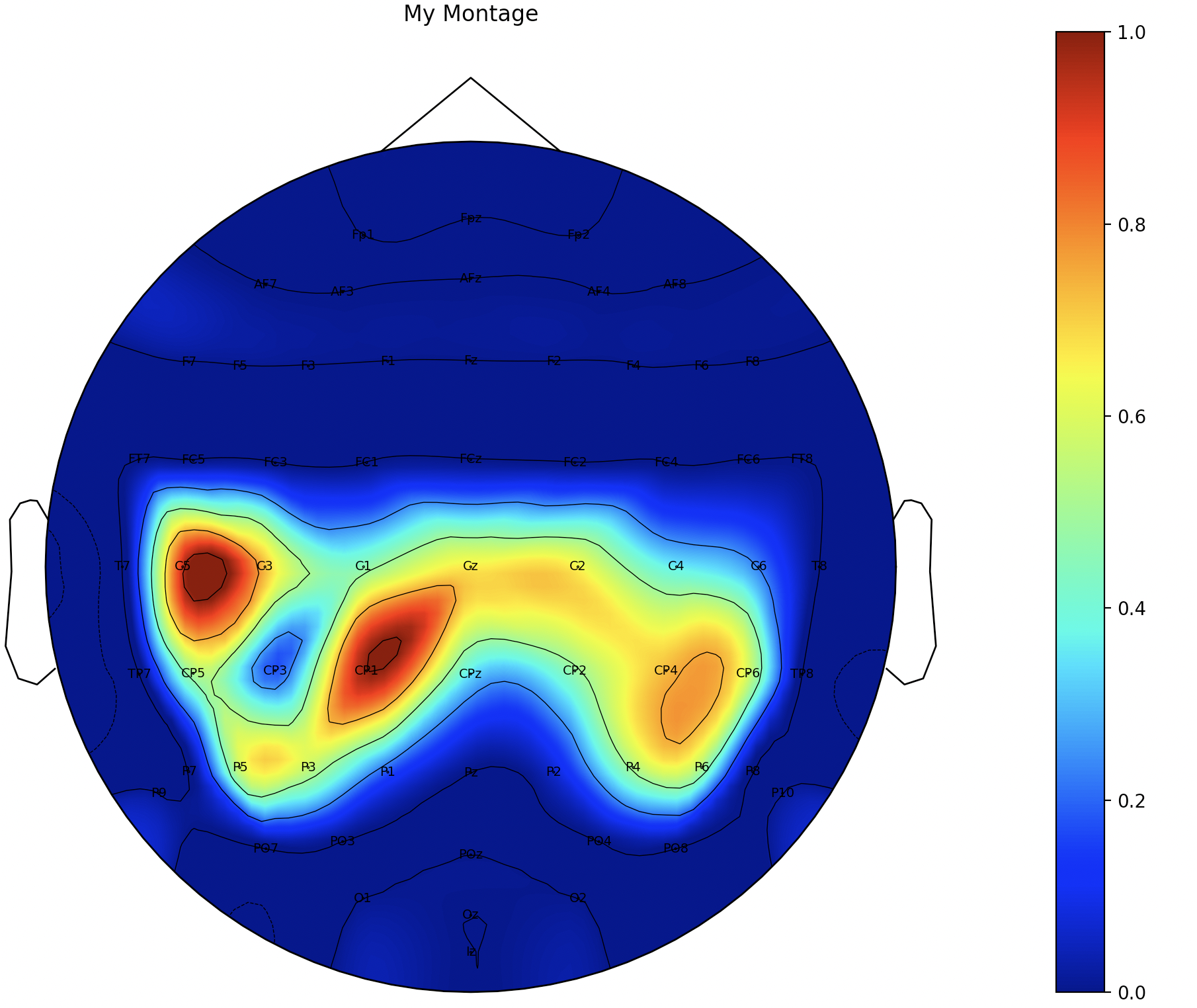
im, cn = mne.viz.plot_topomap(reMyWeight,
myinfo,
names = my_chLa_index,
# vlim=(-2, 2)
)
plt.colorbar(im)
plt.title('My Montage')
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
5、第二种方法
5.1 读取标准的64通道电极布局系统坐标
# 读取MNE中biosemi64电极位置信息
biosemi_montage = mne.channels.make_standard_montage('biosemi64')
sensor_data_64 = biosemi_montage.get_positions()['ch_pos']
sensor_dataframe_64 = pd.DataFrame(sensor_data_64).T
chLa_index_64 = sensor_dataframe_64.index.values
biosemi_montage.plot()
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
5.2 构建自定义部分导联字典
myWeight = {'C1': 0.31, 'C3': 1.22, 'P6': 0.71, 'P4': 0.55,'P2': -1.19, 'CP5': 0.61,
'CP3': -0.80, 'CP1': 2.36, 'P1': -0.74, 'P3': 0.72,'P5': 0.93, 'Pz': -1.46,
'CPz': -0.12, 'Cz': 1.074,'C5':2.34,'C2': 1.04, 'C4': -0.065, 'C6': -0.52,
'CP2': 0.37, 'CP4': 1.30, 'CP6': 0.94}
- 1
- 2
- 3
- 4
5.3 对字典的值进行归一化
norMyWeiht = myWeight.copy()
# Step 1: 获取需要归一化的值
values = [v for v in norMyWeiht.values()]
# Step 2: 找到最小值和最大值
min_value = min(values)
max_value = max(values)
# Step 3: 对值进行归一化计算
for key, value in norMyWeiht.items():
normalized_value = (value - min_value) / (max_value - min_value)
norMyWeiht[key] = normalized_value
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
5.4 根据脑地形图导联顺序重构标准化后的矩阵,将未被选择的导联值置为0
print('脑地形图矩阵导联顺序:',chLa_index_64)
# 重构自定义矩阵顺序
reNorMyWeight = []
for key in chLa_index_64:
if key in norMyWeiht:
val = norMyWeiht[key]
reNorMyWeight.append(val)
else:
reNorMyWeight.append(0)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
5.5 TopoMap可视化
5.5.1 创建info
info = mne.create_info(
ch_names = chLa_index_64.tolist(),
ch_types = ['eeg']*len(reNorMyWeight), # 通道个数
sfreq = 1000) # 采样频率
info.set_montage(biosemi_montage)
- 1
- 2
- 3
- 4
- 5
5.5.2 可视化
im, cn = mne.viz.plot_topomap(reNorMyWeight,
info,
names = chLa_index_64,
cmap = 'jet'
# vlim=(-2, 2)
)
plt.colorbar(im)
plt.title('My Montage')
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
!!!!注意
在加载python库的时候,我添加了:%matplotlib qt 嵌入Python Qt界面
所以出来的图可能是这样的:

需要点击放大:
6、代码下载
https://weisihong9.github.io/2023/07/03/EEG_TopoMap/
下一篇:python绘制局部EEG导联的脑地形图
声明:本文内容由网友自发贡献,不代表【wpsshop博客】立场,版权归原作者所有,本站不承担相应法律责任。如您发现有侵权的内容,请联系我们。转载请注明出处:https://www.wpsshop.cn/w/花生_TL007/article/detail/220171
推荐阅读
相关标签