- 1无需WIFI的投屏神器 爱奇艺电视果4G 来自当贝优选 作者 青春向荣_爱奇遇4g投屏器
- 2Android 启用dataBinding报错:Execution failed for task :app:mergeDebugResources Caused by: java.lang.Null_execution failed for task ':app:generatepackagelis
- 3C语言中动态内存相关的4个函数free、malloc、calloc、realloc,常⻅的动态内存的错误
- 42021-04-06_自定义a*
- 5自媒体内容创作ai写作神器:10款你一定要知道的工具! #科技#学习
- 6Waymo Open Dataset (WOD) 数据集详解(比官网详细版)
- 7华为手机百度云息屏后停止下载_华为智选车载智慧屏评测:像手机一般好用,行车体验更便捷...
- 8小程序VR全景图_小程序实现vr全景图
- 9[WinAPI]通过Windows系统CLSID(GUID)打开系统指定窗口,及部分[上帝模式]命令_26ee0668-a00a-44d7-9371-beb064c98683
- 10ArkTS-组件内转场动画_arkts组件转场动画纵向收缩
食品与疾病关系预测算法赛道-baseline食品与疾病关系预测算法赛道-baseline
赞
踩
★★★ 本文源自AlStudio社区精品项目,【点击此处】查看更多精品内容 >>>
一、赛题背景
电子病历、人工智能、物联网设备、5G技术的快速演化,与大规模的数据资源,成为了数字化医疗的全新支撑。
对于医药公司、医疗机构、科技公司,如何整合全新的技术能力、人才资源、数据资源,为数字化医疗的创新提供更强的动力,成为了共同关切的重要话题。
本次数字医疗算法应用创新大赛是对于这一重要话题的主动探索,旨在最大化利用当前高速发展的算法技术与数据资源,培养数字化医疗人才、激励数字化医疗创新。大赛双赛道分别为食品与疾病关系预测算法赛道和生物共融与数字疗法应用赛道。
1.1 赛题任务
本赛道将提供脱敏后的食物与疾病特征,参赛团队根据主办方提供数据,在高度稀疏数据的场景中,进一步挖掘、融合特征并设计模型,以预测食物与疾病的关系。初赛阶段为二分类问题,分类标签分别为 0(无关)、1(存在正面或负面的影响)。
1.2 赛题数据简介
本次算法赛将提供超过 23.5W 的食物、疾病对应关系及其量化得分,其中食物特征超过 200 个,疾病特征由 3 种不同的方式抽取,累积超过 4000 个特征信息。初赛为 0、1 二分类预测,提供食物、疾病特征,与食物疾病的关系标签。
复赛阶段同时评估 0、1 二分类与相关性评级,在原训练集中增加食物疾病相关性评级的标签数据。
1.2.1 训练集
训练集包括疾病特征数据、食物特征数据(共计 348 种食物)、以及食物疾病关系,用于模型训练:
- 疾病特征集:disease_feature1.csv、disease_feature2.csv、disease_feature3.csv
- 食物特征集:train_food.csv
- 食物疾病关系:train_answer.csv
- 「复赛阶段」食物与疾病关系的相关性评级:semi_train_answer.csv
1.2.2 测试集
初赛测试集分两个阶段(A/B 榜),不提供预测结果,其中:
- 初赛第一阶段 A 榜测试集: 2023 年 2 月 22 日中午 12:00:00— 2023 年 3 月 20 日中午 12:00:00,包括 A 榜阶段食物特征数据(共计 115 种食物)与初赛 A 榜提交样例,用于模型结果验证:
preliminary_a_food.csv
preliminary_a_submit_sample.csv
- 1
- 2
- 初赛第二阶段 B 榜测试集: 2023 年 3 月 20 日中午 12:00:00— 2023 年 3 月 22 日中午 12:00:00,包括 B 榜阶段食物特征数据(共计 116 种食物)与初赛 B 榜提交样例,用于模型结果验证:
preliminary_b_food.csv
preliminary_b_submit_sample.csv
- 1
- 2
- 初赛第二阶段 B 测试集与初赛第一阶段 A 榜测试集分布与规模相同,将于 B 榜提交开始后在赛事主页提供下载,最终初赛排名以初赛第二阶段 B 榜成绩为准。
1.3 字段说明
1.3.1 疾病特征
累计包含 407 种疾病的 4630 种特征信息,三种不同的特征抽取方式将疾病特征划分为三部分特征集,数据高度稀疏。
| 字段名称 | 格式 | 解释说明 | 范围/特征集1 | 范围/特征集2 | 范围/特征集3 |
|---|---|---|---|---|---|
| disease_id | 字符串 | 疾病 id | 共涉及 220 种疾病 | 共涉及 301 种疾病 | 共涉及 392 种疾病 |
| F_x | 浮点型 | 疾病特征值 | F_0 ~F_4629,字段名称不连续,共涉及 996 种疾病特征 | F_0 ~F_4629,字段名称不连续,共涉及 3181 种疾病特征 | F_1 ~F_4627,字段名称不连续,共涉及 1453 种疾病特征 |
1.3.2 食物特征
| 序列 | 字段名称 | 格式 | 解释说明 | 示例 |
|---|---|---|---|---|
| 1 | food_id | 字符串 | 食物 id | food_0 |
| 2~213 | N_x | 浮点型 | 212 种食物特征,字段名称从 N_0 ~N_211 | 0.123 |
1.3.3 食物疾病关系
| 序列 | 字段名称 | 格式 | 解释说明 | 示例 |
|---|---|---|---|---|
| 1 | food_id | 字符串 | 食物 id | food_0 |
| 2 | disease_id | 字符串 | 疾病 id | disease_0 |
| 3 | related | 整型 | 食物与疾病是否相关:0(无关)、1(存在正面或负面的影响) | 0 |
1.4 评测方法
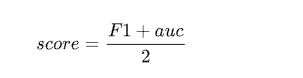
1.5 提交示例
| 序列 | 字段名称 | 格式 | 解释说明 | 示例 |
|---|---|---|---|---|
| 1 | food_id | 字符串 | 食物 id | food_0 |
| 2 | disease_id | 字符串 | 疾病 id | disease_0 |
| 3 | related_prob | 浮点型 | 食品与疾病预测为 1 的概率若 related_prob >= 0.5,评审计算 f1 得分时判定为类别 1 | 0.1 |
1.6 比赛传送门
二、项目介绍
2.1 项目意义
在本次食物与疾病关系预测挑战赛中,参赛团队将获得脱敏后的大量数据集。这些数据集包含了各种类型的食物和相应的健康指标,例如血压、胆固醇等等。这些指标可以帮助我们了解不同种类的食物对人体健康产生的影响。在实际应用中,由于数据量巨大且高度稀疏,传统的特征提取方法难以有效地提取出有用信息。因此,在本次比赛中,参赛团队需要进一步挖掘、融合特征并设计模型,以预测食物与疾病的关系。
在这个 Baseline 中,我们分别尝试复杂的特征结合传统的机器学习模型。我们在解决机器学习问题时,一般会遵循以下流程:
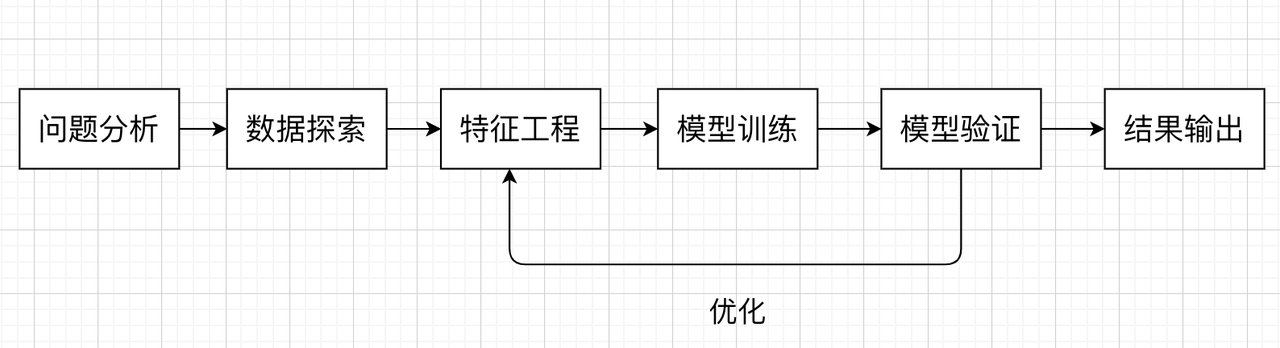
2.2 模型介绍
LightGBM(LightGBM: Gradient Boosting of Big Data Applications)是一种基于梯度提升树模型(Gradient Boosting Decision Tree)的轻量级机器学习算法,用于解决大规模数据集的分类、回归和聚类问题。它的核心思想是利用梯度下降来优化模型参数,从而提高模型的预测准确率。LightGBM的主要特点包括以下几个方面:
- 轻量级:LightGBM采用了基于梯度的优化方法,避免了过多的参数调整,从而减少了计算量和内存消耗。
- 高效性:LightGBM的基本思想是利用梯度下降来不断逼近模型的最优参数,从而减少了搜索空间,提高了模型的效率。
- 可解释性:LightGBM采用了可解释的数据预处理方法,即在训练过程中对数据进行归一化、剪枝等处理,从而提高了模型的可解释性和可靠性。
- 易用性:LightGBM提供了灵活的模型定义方式,可以根据实际需求进行定制化开发,同时也提供了丰富的可视化工具,便于用户理解和操作。
- 通用性:LightGBM可以适用于多种机器学习框架和数据存储格式,具有广泛的应用前景。
- 总之,LightGBM是一种高效、可解释、易用的轻量级机器学习算法,适用于大规模数据集的分类、回归和聚类问题。
三、详细方案实现
3.1 数据分析
3.1.1 数据解压缩
!unzip -d /home/aistudio/data/data200766/ /home/aistudio/data/data200766/初赛A榜测试集.zip
!unzip -d /home/aistudio/data/data200766/ /home/aistudio/data/data200766/初赛B榜测试集.zip
!unzip -d /home/aistudio/data/data200766/ /home/aistudio/data/data200766/训练集.zip
- 1
- 2
- 3
Archive: /home/aistudio/data/data200766/初赛A榜测试集.zip
creating: /home/aistudio/data/data200766/初赛A榜测试集/
inflating: /home/aistudio/data/data200766/初赛A榜测试集/preliminary_a_submit_sample.csv
inflating: /home/aistudio/data/data200766/初赛A榜测试集/preliminary_a_food.csv
Archive: /home/aistudio/data/data200766/初赛B榜测试集.zip
creating: /home/aistudio/data/data200766/初赛B榜测试集/
inflating: /home/aistudio/data/data200766/初赛B榜测试集/preliminary_b_submit_sample.csv
inflating: /home/aistudio/data/data200766/初赛B榜测试集/preliminary_b_food.csv
Archive: /home/aistudio/data/data200766/训练集.zip
creating: /home/aistudio/data/data200766/训练集/
inflating: /home/aistudio/data/data200766/训练集/disease_feature3.csv
inflating: /home/aistudio/data/data200766/训练集/disease_feature2.csv
inflating: /home/aistudio/data/data200766/训练集/disease_feature1.csv
inflating: /home/aistudio/data/data200766/训练集/train_answer.csv
inflating: /home/aistudio/data/data200766/训练集/train_food.csv
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
3.1.2 导入相关库
!pip install --upgrade pip
!git clone --recursive https://github.com/Microsoft/LightGBM
!cd LightGBM && rm -rf build && mkdir build && cd build && cmake -DUSE_GPU=1 ../../LightGBM && make -j4 && cd ../python-package && python3 setup.py install --precompile --gpu;
- 1
- 2
- 3
- 4
- 5
import pandas as pd import os import gc import lightgbm as lgb from sklearn.linear_model import SGDRegressor, LinearRegression, Ridge from sklearn.preprocessing import MinMaxScaler import math import numpy as np from tqdm import tqdm from sklearn.model_selection import StratifiedKFold, KFold, GroupKFold from sklearn.metrics import accuracy_score, f1_score, roc_auc_score, log_loss import matplotlib.pyplot as plt import time import warnings warnings.filterwarnings('ignore')
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
3.1.3 加载数据
disease_feature1 = pd.read_csv("/home/aistudio/data/data200766/训练集/disease_feature1.csv")
disease_feature2 = pd.read_csv("/home/aistudio/data/data200766/训练集/disease_feature2.csv")
disease_feature3 = pd.read_csv("/home/aistudio/data/data200766/训练集/disease_feature3.csv")
train_answer = pd.read_csv("/home/aistudio/data/data200766/训练集/train_answer.csv")
train_food = pd.read_csv("/home/aistudio/data/data200766/训练集/train_food.csv")
preliminary_a_food = pd.read_csv("/home/aistudio/data/data200766/初赛B榜测试集/preliminary_b_food.csv")
preliminary_a_submit_sample = pd.read_csv("/home/aistudio/data/data200766/初赛B榜测试集/preliminary_b_submit_sample.csv")
pd.set_option('display.max_columns', None)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
del preliminary_a_submit_sample['related_prob']
data = pd.concat([train_answer, preliminary_a_submit_sample], axis = 0).reset_index(drop=True)
data.head()
- 1
- 2
- 3
| food_id | disease_id | related | |
|---|---|---|---|
| 0 | food_0 | disease_998 | 0.0 |
| 1 | food_0 | disease_861 | 0.0 |
| 2 | food_0 | disease_559 | 0.0 |
| 3 | food_0 | disease_841 | 0.0 |
| 4 | food_0 | disease_81 | 0.0 |
这里直接使用每个变量后的数字进行编码,当然也可以使用labelencoder的方式。
data['food'] = data['food_id'].apply(lambda x : int(x.split('_')[1]))
data['disease'] = data['disease_id'].apply(lambda x : int(x.split('_')[1]))
- 1
- 2
food = pd.concat([train_food, preliminary_a_food], axis = 0).reset_index(drop=True)
food.head()
- 1
- 2
| food_id | N_0 | N_1 | N_2 | N_3 | N_4 | N_5 | N_6 | N_7 | N_8 | N_9 | N_10 | N_11 | N_12 | N_13 | N_14 | N_15 | N_16 | N_17 | N_18 | N_19 | N_20 | N_21 | N_22 | N_23 | N_24 | N_25 | N_26 | N_27 | N_28 | N_29 | N_30 | N_31 | N_32 | N_33 | N_34 | N_35 | N_36 | N_37 | N_38 | N_39 | N_40 | N_41 | N_42 | N_43 | N_44 | N_45 | N_46 | N_47 | N_48 | N_49 | N_50 | N_51 | N_52 | N_53 | N_54 | N_55 | N_56 | N_57 | N_58 | N_59 | N_60 | N_61 | N_62 | N_63 | N_64 | N_65 | N_66 | N_67 | N_68 | N_69 | N_70 | N_71 | N_72 | N_73 | N_74 | N_75 | N_76 | N_77 | N_78 | N_79 | N_80 | N_81 | N_82 | N_83 | N_84 | N_85 | N_86 | N_87 | N_88 | N_89 | N_90 | N_91 | N_92 | N_93 | N_94 | N_95 | N_96 | N_97 | N_98 | N_99 | N_100 | N_101 | N_102 | N_103 | N_104 | N_105 | N_106 | N_107 | N_108 | N_109 | N_110 | N_111 | N_112 | N_113 | N_114 | N_115 | N_116 | N_117 | N_118 | N_119 | N_120 | N_121 | N_122 | N_123 | N_124 | N_125 | N_126 | N_127 | N_128 | N_129 | N_130 | N_131 | N_132 | N_133 | N_134 | N_135 | N_136 | N_137 | N_138 | N_139 | N_140 | N_141 | N_142 | N_143 | N_144 | N_145 | N_146 | N_147 | N_148 | N_149 | N_150 | N_151 | N_152 | N_153 | N_154 | N_155 | N_156 | N_157 | N_158 | N_159 | N_160 | N_161 | N_162 | N_163 | N_164 | N_165 | N_166 | N_167 | N_168 | N_169 | N_170 | N_171 | N_172 | N_173 | N_174 | N_175 | N_176 | N_177 | N_178 | N_179 | N_180 | N_181 | N_182 | N_183 | N_184 | N_185 | N_186 | N_187 | N_188 | N_189 | N_190 | N_191 | N_192 | N_193 | N_194 | N_195 | N_196 | N_197 | N_198 | N_199 | N_200 | N_201 | N_202 | N_203 | N_204 | N_205 | N_206 | N_207 | N_208 | N_209 | N_210 | N_211 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | food_0 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 32.0 | NaN | NaN | 2.10 | NaN | 6.0 | 87.0 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 14.4 | NaN | NaN | NaN | NaN | 0.157 | NaN | 6.0 | NaN | NaN | NaN | NaN | NaN | NaN | 23.0 | NaN | NaN | NaN | NaN | NaN | 0.056 | 0.409 | 0.069 | NaN | NaN | NaN | NaN | NaN | NaN | 1.9 | NaN | 36.0 | 36.0 | 36.0 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.96 | NaN | NaN | NaN | NaN | 0.0 | 0.0 | NaN | 27.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.000 | NaN | NaN | NaN | 0.056 | NaN | 0.000 | NaN | 0.0 | NaN | NaN | 0.481 | NaN | NaN | NaN | NaN | 70.0 | NaN | NaN | NaN | NaN | 79.0 | NaN | 3.99 | 0.234 | NaN | NaN | NaN | NaN | 0.175 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | NaN | NaN | 0.0 | 0.126 | 0.6 | NaN | 0.0 | NaN | 0.000 | NaN | 0.002 | NaN | 0.059 | NaN | 0.008 | NaN | NaN | NaN | NaN | 0.000 | 0.0 | 0.000 | 6.0 | NaN | NaN | NaN | 0.20 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.076 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.69 | NaN | NaN | NaN | NaN | NaN | 8.0 | 0.0 | 0.0 | 0.034 | 8.2 | 0.0 | NaN | NaN | NaN | NaN | 0.02 | 0.0 | NaN | NaN | 30.5 | 92.82 | NaN | 0.92 |
| 1 | food_1 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 268.0 | NaN | NaN | 21.01 | NaN | 0.0 | 1.0 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 52.1 | NaN | NaN | NaN | NaN | 1.099 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 598.0 | NaN | NaN | NaN | NaN | NaN | 33.076 | 12.955 | 4.092 | NaN | NaN | NaN | NaN | NaN | NaN | 10.9 | NaN | 55.0 | 55.0 | 55.0 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 3.73 | NaN | NaN | NaN | NaN | 1.0 | 0.0 | NaN | 279.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.259 | NaN | NaN | NaN | 32.754 | NaN | 0.007 | NaN | 0.0 | NaN | NaN | 3.637 | NaN | NaN | NaN | NaN | 471.0 | NaN | NaN | NaN | NaN | 713.0 | NaN | 20.96 | 12.945 | NaN | NaN | NaN | NaN | 0.010 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | NaN | NaN | 0.0 | 1.197 | 2.0 | NaN | 0.0 | NaN | 0.000 | NaN | 0.019 | NaN | 3.348 | NaN | 0.704 | NaN | NaN | NaN | NaN | 0.000 | 0.0 | 0.000 | 3.0 | NaN | NaN | NaN | 4.86 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.077 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 52.54 | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.0 | 0.0 | 0.136 | 0.0 | 0.0 | NaN | NaN | NaN | NaN | 23.90 | 0.0 | NaN | NaN | 0.0 | 2.41 | NaN | 3.31 |
| 2 | food_4 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 62.0 | NaN | NaN | 79.32 | NaN | 0.0 | 0.0 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 11.1 | NaN | NaN | NaN | NaN | 0.272 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 299.0 | NaN | NaN | NaN | NaN | NaN | 0.024 | 0.053 | 0.094 | NaN | NaN | NaN | NaN | NaN | NaN | 4.5 | NaN | 5.0 | 5.0 | 5.0 | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.79 | NaN | NaN | NaN | NaN | 0.0 | 0.0 | NaN | 36.0 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.001 | NaN | NaN | NaN | 0.023 | NaN | 0.000 | NaN | 0.0 | NaN | NaN | 0.766 | NaN | NaN | NaN | NaN | 98.0 | NaN | NaN | NaN | NaN | 744.0 | NaN | 3.30 | 0.039 | NaN | NaN | NaN | NaN | 0.014 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | NaN | NaN | 0.0 | 0.125 | 0.6 | NaN | 0.0 | NaN | 0.001 | NaN | 0.004 | NaN | 0.056 | NaN | 0.013 | NaN | NaN | NaN | NaN | 0.007 | 0.0 | 0.001 | 26.0 | NaN | NaN | NaN | 65.18 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.106 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.25 | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.0 | 0.0 | 0.174 | 2.3 | 0.0 | NaN | NaN | NaN | NaN | 0.12 | 0.0 | NaN | NaN | 3.5 | 15.46 | NaN | 0.36 |
| 3 | food_5 | NaN | NaN | NaN | 0.068 | 0.0 | 0.045 | 0.75 | 0.314 | NaN | NaN | NaN | NaN | NaN | 0.0 | 13.0 | NaN | NaN | 11.12 | NaN | 19.0 | 1094.0 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 2.8 | NaN | NaN | NaN | NaN | 0.078 | NaN | 104.0 | NaN | 0.003 | NaN | NaN | NaN | NaN | 124.5 | NaN | NaN | NaN | NaN | NaN | 0.170 | 0.077 | 0.027 | 0.0 | NaN | NaN | NaN | NaN | NaN | 2.0 | NaN | 9.0 | 9.0 | 9.0 | 0.0 | 0.94 | 0.0 | NaN | NaN | 2.37 | 0.157 | 0.040 | NaN | 0.027 | NaN | NaN | 0.39 | 0.041 | 0.0 | 0.077 | NaN | 89.0 | 0.0 | 0.097 | 10.0 | NaN | 0.06 | 0.077 | 0.006 | NaN | NaN | NaN | 0.000 | NaN | NaN | NaN | 0.170 | NaN | 0.000 | NaN | 0.0 | NaN | NaN | 0.600 | NaN | NaN | 0.240 | 0.052 | 23.0 | NaN | NaN | 18.0 | NaN | 259.0 | 0.101 | 1.40 | 0.077 | NaN | NaN | NaN | NaN | 0.000 | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | NaN | NaN | 0.0 | 0.040 | 0.1 | 0.083 | 0.0 | NaN | 0.000 | NaN | 0.000 | NaN | 0.024 | NaN | 0.003 | NaN | NaN | NaN | NaN | 0.000 | 0.0 | 0.000 | 1.0 | NaN | NaN | 5.87 | 9.24 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.030 | 0.047 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.39 | NaN | 0.015 | 0.029 | 0.047 | 1926.0 | 96.0 | 0.0 | 0.0 | 0.054 | 10.0 | 0.0 | 0.0 | NaN | NaN | NaN | 0.89 | 0.0 | NaN | NaN | 3.3 | 86.35 | NaN | 0.20 |
| 4 | food_6 | NaN | NaN | NaN | 0.115 | 0.0 | 0.091 | 0.58 | 0.508 | NaN | NaN | 0.6 | NaN | NaN | 0.0 | 24.0 | NaN | NaN | 3.88 | NaN | 9.0 | 449.0 | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 16.0 | NaN | NaN | NaN | NaN | 0.189 | NaN | 0.0 | NaN | 0.031 | NaN | NaN | NaN | NaN | 52.5 | NaN | NaN | NaN | NaN | NaN | 0.000 | 0.050 | 0.040 | 0.0 | NaN | NaN | NaN | NaN | NaN | 2.1 | NaN | 52.0 | 52.0 | 52.0 | 0.0 | 1.00 | 0.0 | NaN | NaN | 0.65 | 0.233 | 0.093 | NaN | 0.049 | NaN | NaN | 2.14 | 0.075 | 0.0 | 0.128 | NaN | 710.0 | 0.0 | 0.104 | 14.0 | NaN | 0.00 | 0.158 | 0.031 | 0.0 | NaN | 0.0 | 0.000 | NaN | 0.0 | NaN | 0.000 | NaN | 0.000 | NaN | 0.0 | NaN | NaN | 0.978 | NaN | NaN | 0.274 | 0.075 | 52.0 | NaN | NaN | 24.0 | NaN | 202.0 | 0.071 | 2.20 | 0.040 | NaN | NaN | NaN | NaN | 0.010 | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | 0.0 | NaN | NaN | NaN | 0.0 | NaN | NaN | 0.0 | NaN | NaN | NaN | NaN | NaN | 0.0 | NaN | 0.0 | NaN | NaN | 0.0 | 0.141 | 2.3 | 0.106 | 0.0 | NaN | 0.000 | NaN | 0.000 | 0.0 | 0.040 | 0.0 | 0.000 | 0.0 | NaN | 0.0 | 0.0 | 0.000 | 0.0 | 0.000 | 2.0 | NaN | NaN | 0.23 | 1.88 | NaN | NaN | NaN | NaN | NaN | NaN | 0.0 | 0.143 | 0.084 | 0.0 | 0.0 | 0.09 | NaN | NaN | NaN | NaN | NaN | NaN | 0.12 | NaN | 0.027 | 0.052 | 0.115 | 756.0 | 38.0 | 0.0 | 0.0 | 0.091 | 5.6 | 0.0 | 0.0 | NaN | NaN | NaN | 1.13 | 0.0 | 0.0 | NaN | 41.6 | 93.22 | NaN | 0.54 |
3.1.4 EDA
# 查看数据缺失情况
pd.set_option('display.max_rows', None)
((food.isnull().sum())/food.shape[0]).sort_values(ascending=False).map(lambda x:"{:.2%}".format(x))
- 1
- 2
- 3
- 4

#只保留缺失率少于10%的列
food=food[['N_198','N_33','N_211','N_82','N_101','N_42','N_111','N_165','N_177','N_146','N_17','N_113','N_106','N_14','N_74','N_209','N_188','food_id' ]]
food.head(5)
- 1
- 2
- 3
| N_198 | N_33 | N_211 | N_82 | N_101 | N_42 | N_111 | N_165 | N_177 | N_146 | N_17 | N_113 | N_106 | N_14 | N_74 | N_209 | N_188 | food_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.10 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | food_0 |
| 1 | 0.0 | 1.099 | 3.31 | 279.0 | 3.637 | 598.0 | 713.0 | 3.0 | 0.077 | 1.197 | 21.01 | 20.96 | 471.0 | 268.0 | 3.73 | 2.41 | 52.54 | food_1 |
| 2 | 2.3 | 0.272 | 0.36 | 36.0 | 0.766 | 299.0 | 744.0 | 26.0 | 0.106 | 0.125 | 79.32 | 3.30 | 98.0 | 62.0 | 1.79 | 15.46 | 0.25 | food_4 |
| 3 | 10.0 | 0.078 | 0.20 | 10.0 | 0.600 | 124.5 | 259.0 | 1.0 | 0.030 | 0.040 | 11.12 | 1.40 | 23.0 | 13.0 | 0.39 | 86.35 | 0.39 | food_5 |
| 4 | 5.6 | 0.189 | 0.54 | 14.0 | 0.978 | 52.5 | 202.0 | 2.0 | 0.143 | 0.141 | 3.88 | 2.20 | 52.0 | 24.0 | 2.14 | 93.22 | 0.12 | food_6 |
3.2 数据处理
3.2.1 目标编码
由于本题只有两个离散变量food_id 和disease_id ,而测试集中都是新的foodid
cat_list = ['disease'] def stat(df, df_merge, group_by, agg): group = df.groupby(group_by).agg(agg) columns = [] for on, methods in agg.items(): for method in methods: columns.append('{}_{}_{}'.format('_'.join(group_by), on, method)) group.columns = columns group.reset_index(inplace=True) df_merge = df_merge.merge(group, on=group_by, how='left') del (group) gc.collect() return df_merge def statis_feat(df_know, df_unknow,cat_list): for f in tqdm(cat_list): df_unknow = stat(df_know, df_unknow, [f], {'related': ['mean']}) return df_unknow df_train = data[~data['related'].isnull()] df_train = df_train.reset_index(drop=True) df_test = data[data['related'].isnull()] df_stas_feat = None kf = StratifiedKFold(n_splits=5, random_state=2020, shuffle=True) for train_index, val_index in kf.split(df_train, df_train['related']): df_fold_train = df_train.iloc[train_index] df_fold_val = df_train.iloc[val_index] df_fold_val = statis_feat(df_fold_train, df_fold_val,cat_list) df_stas_feat = pd.concat([df_stas_feat, df_fold_val], axis=0) del (df_fold_train) del (df_fold_val) gc.collect() df_test = statis_feat(df_train, df_test,cat_list) data = pd.concat([df_stas_feat, df_test], axis=0) data = data.reset_index(drop=True) del (df_stas_feat) del (df_train) del (df_test)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
100%|██████████| 1/1 [00:00<00:00, 7.57it/s]
100%|██████████| 1/1 [00:00<00:00, 9.00it/s]
100%|██████████| 1/1 [00:00<00:00, 9.45it/s]
100%|██████████| 1/1 [00:00<00:00, 8.77it/s]
100%|██████████| 1/1 [00:00<00:00, 8.65it/s]
100%|██████████| 1/1 [00:00<00:00, 9.16it/s]
- 1
- 2
- 3
- 4
- 5
- 6
3.2.1 疾病特征处理
使用TruncatedSVD 的方法,对疾病特征进行降维,维度均为128
f_col = [col for col in disease_feature1.columns if 'F' in col]
- 1
disease_feature_1_ = disease_feature1.copy()
from sklearn.feature_extraction.text import CountVectorizer,TfidfVectorizer,TfidfTransformer
from sklearn.decomposition import TruncatedSVD, SparsePCA
disease_feature_1_ = disease_feature_1_.fillna(0)
decom=TruncatedSVD(n_components=128, n_iter = 20, random_state=2023)
decom_x=decom.fit_transform(disease_feature_1_.iloc[:,1:])
decom_feas=pd.DataFrame(decom_x)
decom_feas.columns=['disease1_svd_'+str(i) for i in range(decom_x.shape[1])]
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
disease_feature1 = disease_feature1[['disease_id']]
for col in decom_feas:
disease_feature1[col] = decom_feas[col]
- 1
- 2
- 3
f_col = [col for col in disease_feature2.columns if 'F' in col]
- 1
disease_feature_2_ = disease_feature2.copy()
from sklearn.feature_extraction.text import CountVectorizer,TfidfVectorizer,TfidfTransformer
from sklearn.decomposition import TruncatedSVD, SparsePCA
disease_feature_2_ = disease_feature_2_.fillna(0)
decom=TruncatedSVD(n_components=128, n_iter = 20, random_state=2023)
decom_x=decom.fit_transform(disease_feature_2_.iloc[:,1:])
decom_feas=pd.DataFrame(decom_x)
decom_feas.columns=['disease2_svd_'+str(i) for i in range(decom_x.shape[1])]
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
disease_feature2 = disease_feature2[['disease_id']]
for col in decom_feas:
disease_feature2[col] = decom_feas[col]
- 1
- 2
- 3
f_col = [col for col in disease_feature3.columns if 'F' in col]
- 1
disease_feature_3_ = disease_feature3.copy()
from sklearn.feature_extraction.text import CountVectorizer,TfidfVectorizer,TfidfTransformer
from sklearn.decomposition import TruncatedSVD, SparsePCA
disease_feature_3_ = disease_feature_3_.fillna(0)
decom=TruncatedSVD(n_components=128, n_iter = 20, random_state=2023)
decom_x=decom.fit_transform(disease_feature_3_.iloc[:,1:])
decom_feas=pd.DataFrame(decom_x)
decom_feas.columns=['disease3_svd_'+str(i) for i in range(decom_x.shape[1])]
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
disease_feature3 = disease_feature3[['disease_id']]
for col in decom_feas:
disease_feature3[col] = decom_feas[col]
- 1
- 2
- 3
data = data.merge(food, on = 'food_id', how = 'left')
data = data.merge(disease_feature1, on = 'disease_id', how = 'left')
data = data.merge(disease_feature2, on = 'disease_id', how = 'left')
data = data.merge(disease_feature3, on = 'disease_id', how = 'left')
data.head()
- 1
- 2
- 3
- 4
- 5
| food_id | disease_id | related | food | disease | disease_related_mean | N_198 | N_33 | N_211 | N_82 | N_101 | N_42 | N_111 | N_165 | N_177 | N_146 | N_17 | N_113 | N_106 | N_14 | N_74 | N_209 | N_188 | disease1_svd_0 | disease1_svd_1 | disease1_svd_2 | disease1_svd_3 | disease1_svd_4 | disease1_svd_5 | disease1_svd_6 | disease1_svd_7 | disease1_svd_8 | disease1_svd_9 | disease1_svd_10 | disease1_svd_11 | disease1_svd_12 | disease1_svd_13 | disease1_svd_14 | disease1_svd_15 | disease1_svd_16 | disease1_svd_17 | disease1_svd_18 | disease1_svd_19 | disease1_svd_20 | disease1_svd_21 | disease1_svd_22 | disease1_svd_23 | disease1_svd_24 | disease1_svd_25 | disease1_svd_26 | disease1_svd_27 | disease1_svd_28 | disease1_svd_29 | disease1_svd_30 | disease1_svd_31 | disease1_svd_32 | disease1_svd_33 | disease1_svd_34 | disease1_svd_35 | disease1_svd_36 | disease1_svd_37 | disease1_svd_38 | disease1_svd_39 | disease1_svd_40 | disease1_svd_41 | disease1_svd_42 | disease1_svd_43 | disease1_svd_44 | disease1_svd_45 | disease1_svd_46 | disease1_svd_47 | disease1_svd_48 | disease1_svd_49 | disease1_svd_50 | disease1_svd_51 | disease1_svd_52 | disease1_svd_53 | disease1_svd_54 | disease1_svd_55 | disease1_svd_56 | disease1_svd_57 | disease1_svd_58 | disease1_svd_59 | disease1_svd_60 | disease1_svd_61 | disease1_svd_62 | disease1_svd_63 | disease1_svd_64 | disease1_svd_65 | disease1_svd_66 | disease1_svd_67 | disease1_svd_68 | disease1_svd_69 | disease1_svd_70 | disease1_svd_71 | disease1_svd_72 | disease1_svd_73 | disease1_svd_74 | disease1_svd_75 | disease1_svd_76 | disease1_svd_77 | disease1_svd_78 | disease1_svd_79 | disease1_svd_80 | disease1_svd_81 | disease1_svd_82 | disease1_svd_83 | disease1_svd_84 | disease1_svd_85 | disease1_svd_86 | disease1_svd_87 | disease1_svd_88 | disease1_svd_89 | disease1_svd_90 | disease1_svd_91 | disease1_svd_92 | disease1_svd_93 | disease1_svd_94 | disease1_svd_95 | disease1_svd_96 | disease1_svd_97 | disease1_svd_98 | disease1_svd_99 | disease1_svd_100 | disease1_svd_101 | disease1_svd_102 | disease1_svd_103 | disease1_svd_104 | disease1_svd_105 | disease1_svd_106 | disease1_svd_107 | disease1_svd_108 | disease1_svd_109 | disease1_svd_110 | disease1_svd_111 | disease1_svd_112 | disease1_svd_113 | disease1_svd_114 | disease1_svd_115 | disease1_svd_116 | disease1_svd_117 | disease1_svd_118 | disease1_svd_119 | disease1_svd_120 | disease1_svd_121 | disease1_svd_122 | disease1_svd_123 | disease1_svd_124 | disease1_svd_125 | disease1_svd_126 | disease1_svd_127 | disease2_svd_0 | disease2_svd_1 | disease2_svd_2 | disease2_svd_3 | disease2_svd_4 | disease2_svd_5 | disease2_svd_6 | disease2_svd_7 | disease2_svd_8 | disease2_svd_9 | disease2_svd_10 | disease2_svd_11 | disease2_svd_12 | disease2_svd_13 | disease2_svd_14 | disease2_svd_15 | disease2_svd_16 | disease2_svd_17 | disease2_svd_18 | disease2_svd_19 | disease2_svd_20 | disease2_svd_21 | disease2_svd_22 | disease2_svd_23 | disease2_svd_24 | disease2_svd_25 | disease2_svd_26 | disease2_svd_27 | disease2_svd_28 | disease2_svd_29 | disease2_svd_30 | disease2_svd_31 | disease2_svd_32 | disease2_svd_33 | disease2_svd_34 | disease2_svd_35 | disease2_svd_36 | disease2_svd_37 | disease2_svd_38 | disease2_svd_39 | disease2_svd_40 | disease2_svd_41 | disease2_svd_42 | disease2_svd_43 | disease2_svd_44 | disease2_svd_45 | disease2_svd_46 | disease2_svd_47 | disease2_svd_48 | disease2_svd_49 | disease2_svd_50 | disease2_svd_51 | disease2_svd_52 | disease2_svd_53 | disease2_svd_54 | disease2_svd_55 | disease2_svd_56 | disease2_svd_57 | disease2_svd_58 | disease2_svd_59 | disease2_svd_60 | disease2_svd_61 | disease2_svd_62 | disease2_svd_63 | disease2_svd_64 | disease2_svd_65 | disease2_svd_66 | disease2_svd_67 | disease2_svd_68 | disease2_svd_69 | disease2_svd_70 | disease2_svd_71 | disease2_svd_72 | disease2_svd_73 | disease2_svd_74 | disease2_svd_75 | disease2_svd_76 | disease2_svd_77 | disease2_svd_78 | disease2_svd_79 | disease2_svd_80 | disease2_svd_81 | disease2_svd_82 | disease2_svd_83 | disease2_svd_84 | disease2_svd_85 | disease2_svd_86 | disease2_svd_87 | disease2_svd_88 | disease2_svd_89 | disease2_svd_90 | disease2_svd_91 | disease2_svd_92 | disease2_svd_93 | disease2_svd_94 | disease2_svd_95 | disease2_svd_96 | disease2_svd_97 | disease2_svd_98 | disease2_svd_99 | disease2_svd_100 | disease2_svd_101 | disease2_svd_102 | disease2_svd_103 | disease2_svd_104 | disease2_svd_105 | disease2_svd_106 | disease2_svd_107 | disease2_svd_108 | disease2_svd_109 | disease2_svd_110 | disease2_svd_111 | disease2_svd_112 | disease2_svd_113 | disease2_svd_114 | disease2_svd_115 | disease2_svd_116 | disease2_svd_117 | disease2_svd_118 | disease2_svd_119 | disease2_svd_120 | disease2_svd_121 | disease2_svd_122 | disease2_svd_123 | disease2_svd_124 | disease2_svd_125 | disease2_svd_126 | disease2_svd_127 | disease3_svd_0 | disease3_svd_1 | disease3_svd_2 | disease3_svd_3 | disease3_svd_4 | disease3_svd_5 | disease3_svd_6 | disease3_svd_7 | disease3_svd_8 | disease3_svd_9 | disease3_svd_10 | disease3_svd_11 | disease3_svd_12 | disease3_svd_13 | disease3_svd_14 | disease3_svd_15 | disease3_svd_16 | disease3_svd_17 | disease3_svd_18 | disease3_svd_19 | disease3_svd_20 | disease3_svd_21 | disease3_svd_22 | disease3_svd_23 | disease3_svd_24 | disease3_svd_25 | disease3_svd_26 | disease3_svd_27 | disease3_svd_28 | disease3_svd_29 | disease3_svd_30 | disease3_svd_31 | disease3_svd_32 | disease3_svd_33 | disease3_svd_34 | disease3_svd_35 | disease3_svd_36 | disease3_svd_37 | disease3_svd_38 | disease3_svd_39 | disease3_svd_40 | disease3_svd_41 | disease3_svd_42 | disease3_svd_43 | disease3_svd_44 | disease3_svd_45 | disease3_svd_46 | disease3_svd_47 | disease3_svd_48 | disease3_svd_49 | disease3_svd_50 | disease3_svd_51 | disease3_svd_52 | disease3_svd_53 | disease3_svd_54 | disease3_svd_55 | disease3_svd_56 | disease3_svd_57 | disease3_svd_58 | disease3_svd_59 | disease3_svd_60 | disease3_svd_61 | disease3_svd_62 | disease3_svd_63 | disease3_svd_64 | disease3_svd_65 | disease3_svd_66 | disease3_svd_67 | disease3_svd_68 | disease3_svd_69 | disease3_svd_70 | disease3_svd_71 | disease3_svd_72 | disease3_svd_73 | disease3_svd_74 | disease3_svd_75 | disease3_svd_76 | disease3_svd_77 | disease3_svd_78 | disease3_svd_79 | disease3_svd_80 | disease3_svd_81 | disease3_svd_82 | disease3_svd_83 | disease3_svd_84 | disease3_svd_85 | disease3_svd_86 | disease3_svd_87 | disease3_svd_88 | disease3_svd_89 | disease3_svd_90 | disease3_svd_91 | disease3_svd_92 | disease3_svd_93 | disease3_svd_94 | disease3_svd_95 | disease3_svd_96 | disease3_svd_97 | disease3_svd_98 | disease3_svd_99 | disease3_svd_100 | disease3_svd_101 | disease3_svd_102 | disease3_svd_103 | disease3_svd_104 | disease3_svd_105 | disease3_svd_106 | disease3_svd_107 | disease3_svd_108 | disease3_svd_109 | disease3_svd_110 | disease3_svd_111 | disease3_svd_112 | disease3_svd_113 | disease3_svd_114 | disease3_svd_115 | disease3_svd_116 | disease3_svd_117 | disease3_svd_118 | disease3_svd_119 | disease3_svd_120 | disease3_svd_121 | disease3_svd_122 | disease3_svd_123 | disease3_svd_124 | disease3_svd_125 | disease3_svd_126 | disease3_svd_127 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | food_0 | disease_861 | 0.0 | 0 | 861 | 0.003521 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.1 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 21.207893 | 0.450860 | 0.247976 | -0.094030 | -0.010093 | -0.165738 | 0.063872 | -0.106507 | -0.206626 | 0.123055 | -0.046291 | 0.103308 | 0.114410 | -0.150404 | 0.008024 | -0.075472 | 0.328313 | -0.054091 | 0.210355 | 0.139096 | -0.028611 | 0.080221 | -0.151398 | 0.034177 | 0.041904 | 0.002051 | -0.131626 | 0.076600 | -0.131167 | 0.006474 | -0.097548 | 0.012424 | 0.028256 | 0.006985 | 0.010723 | 0.017358 | -0.011925 | -0.021033 | 0.015393 | 0.006445 | -0.019345 | -0.033685 | -0.030142 | 0.020834 | 0.023872 | -0.025286 | 0.003081 | 0.007100 | 0.019440 | 0.003406 | -0.015664 | 0.007384 | 0.004627 | 0.010140 | 0.016964 | -0.001596 | -0.000890 | -0.002959 | 0.004079 | -0.001305 | 0.000426 | 0.000269 | 0.000391 | 0.000193 | 1.491195e-04 | -0.000022 | 0.000075 | -0.000328 | -0.000104 | -0.000818 | -0.000536 | 0.000425 | 0.000675 | 0.000525 | -0.000909 | 0.000450 | -0.000754 | 0.002798 | -0.001876 | -0.003088 | -0.002775 | -0.001081 | 0.000486 | 0.004900 | 0.000529 | 0.004128 | 0.009350 | 0.002341 | -0.007186 | -0.009234 | -0.007480 | 0.001355 | 0.003528 | -0.017621 | 0.005953 | 0.010327 | 0.004300 | 0.024545 | 0.016885 | 0.013065 | -0.004008 | 0.020067 | -0.027640 | -0.020863 | 0.002466 | -0.009659 | -0.004832 | -0.006345 | 0.007009 | 0.018090 | -0.005579 | -0.001987 | 0.037768 | 0.012893 | 0.002925 | 0.044773 | -0.038541 | 0.005572 | 0.015253 | 0.031836 | -0.001107 | 0.059203 | 0.009907 | -0.044329 | 0.045791 | -0.019859 | -0.008714 | -0.057113 | 1.081144 | -0.096631 | 0.610973 | 0.214094 | 0.285724 | 0.037680 | 2.024824 | 0.396048 | -0.701736 | -0.903993 | -0.163829 | -0.665074 | -0.146170 | 0.292248 | -0.264666 | 0.232114 | 0.142807 | 0.018816 | 0.006140 | 0.215335 | 0.123988 | -0.007711 | 0.503743 | 0.082692 | -0.192049 | 0.049991 | 0.284193 | -0.419728 | -0.265826 | 0.219807 | -0.083213 | 0.365347 | 0.014522 | 0.270022 | -0.100975 | -0.027756 | -0.005296 | -0.384114 | -0.131914 | -0.386147 | 0.037262 | 0.123429 | -0.193423 | 0.233849 | -0.135330 | 0.132876 | -0.204344 | 0.048369 | -0.094278 | -0.219713 | -0.040830 | -0.112828 | -0.003695 | -0.135492 | 0.027470 | -0.069589 | -0.110494 | 0.046717 | -0.214439 | 0.046134 | 0.134354 | -0.104612 | -0.104119 | -0.080847 | 0.155709 | 0.017000 | -0.169362 | 0.020168 | 0.120298 | 0.094487 | -0.117866 | 0.041941 | -0.236982 | -0.036641 | 0.017555 | 0.030866 | 0.045257 | 0.070255 | -0.035509 | 0.031680 | -0.041112 | -0.003302 | -0.026433 | 0.150842 | -0.117318 | -0.081397 | 0.013877 | -0.030950 | 0.035688 | -0.022059 | -0.015251 | -0.046003 | -0.003584 | 0.058973 | 0.116162 | -0.102288 | -0.026451 | -0.065339 | -0.016272 | -0.020509 | 0.050898 | -0.088068 | 0.030054 | -0.022857 | 0.045037 | -0.100831 | 0.004140 | 0.012740 | -0.070383 | 0.056177 | -0.029545 | -0.034183 | 0.027649 | 0.003119 | 0.028489 | -0.058005 | 0.060475 | 0.159239 | 0.003554 | -0.050520 | -0.049823 | -0.017363 | -0.116130 | 0.085801 | 0.072854 | 0.120381 | 0.033087 | -0.025249 |
| 1 | food_0 | disease_839 | 0.0 | 0 | 839 | 0.007299 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.1 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 35.865549 | -0.229259 | -0.043399 | -0.001870 | -0.052036 | -0.012363 | -0.024822 | -0.016792 | -0.026163 | -0.001283 | -0.001989 | 0.002888 | -0.007876 | 0.008097 | 0.001597 | -0.000355 | -0.000594 | -0.002079 | 0.020368 | -0.003079 | 0.021870 | -0.008940 | 0.004237 | -0.004279 | 0.015770 | 0.001230 | 0.004035 | -0.001987 | 0.010776 | -0.003411 | -0.009880 | 0.001792 | -0.013636 | -0.000586 | 0.011546 | -0.014159 | 0.000428 | -0.002775 | -0.001416 | 0.009633 | 0.003450 | -0.003883 | -0.002826 | 0.000251 | 0.009810 | -0.008360 | -0.000637 | 0.003437 | -0.002244 | 0.002247 | -0.000411 | 0.000358 | 0.000566 | 0.002944 | -0.000523 | -0.000021 | -0.000101 | -0.000175 | 0.000425 | 0.000068 | 0.000147 | 0.000108 | -0.000043 | -0.000021 | 1.726565e-05 | -0.000090 | -0.000079 | 0.000011 | -0.000041 | 0.000142 | -0.000004 | 0.000041 | -0.000045 | -0.000070 | -0.000194 | 0.000516 | 0.000210 | 0.000677 | -0.000440 | -0.000186 | -0.000202 | -0.000044 | -0.000397 | 0.001779 | 0.000118 | -0.000687 | 0.000381 | 0.000223 | 0.000390 | -0.000753 | -0.000451 | -0.002448 | -0.001939 | -0.001105 | -0.000197 | -0.001683 | 0.004916 | 0.001206 | -0.000385 | -0.004373 | 0.004536 | 0.001937 | -0.003012 | -0.001530 | 0.002150 | 0.001011 | -0.001764 | 0.003546 | -0.001815 | 0.002938 | -0.003030 | -0.009093 | -0.002076 | -0.004467 | 0.007585 | 0.001704 | -0.000898 | -0.000604 | -0.002589 | 0.007437 | 0.006040 | 0.009314 | 0.007324 | -0.008239 | -0.007265 | -0.007121 | -0.002937 | -0.000852 | 1.817120 | -1.784037 | -0.973115 | -0.322312 | 0.991550 | 2.330509 | -0.176316 | -0.130993 | -0.397610 | -0.210186 | 0.532973 | 0.323650 | 0.015021 | 0.053810 | 0.220851 | -0.044089 | -0.129121 | 0.357842 | -0.118835 | 0.533157 | 0.197675 | 0.033118 | 0.212645 | 0.058697 | -0.138553 | -0.074376 | -0.076036 | 0.418851 | -0.044005 | -0.103922 | 0.011587 | -0.067979 | -0.088224 | -0.190870 | 0.234372 | 0.014150 | 0.295512 | 0.208233 | -0.002893 | 0.028525 | 0.142613 | -0.235298 | 0.071875 | 0.035765 | -0.018080 | 0.006652 | 0.144174 | 0.107761 | 0.102500 | -0.028223 | -0.166622 | -0.227230 | 0.059775 | -0.054151 | 0.143829 | 0.048532 | 0.072578 | -0.095288 | -0.048502 | 0.015662 | 0.030757 | -0.050883 | 0.147684 | 0.008403 | -0.135227 | -0.081102 | -0.040909 | -0.055877 | 0.008944 | 0.042602 | -0.086106 | -0.070377 | -0.061552 | 0.005882 | -0.013598 | 0.051109 | -0.037114 | 0.007101 | -0.044109 | 0.004390 | 0.032941 | -0.026301 | -0.014711 | 0.021407 | 0.017150 | 0.007859 | -0.012711 | -0.081762 | 0.013885 | -0.030356 | -0.024276 | -0.090971 | 0.031763 | -0.072141 | 0.062488 | 0.036716 | -0.080577 | -0.054123 | 0.011628 | -0.033164 | 0.075565 | 0.054723 | 0.017667 | 0.054461 | 0.015337 | -0.044796 | -0.028650 | -0.086811 | -0.008269 | 0.086882 | -0.057133 | 0.054998 | -0.053585 | -0.027943 | -0.031830 | 0.060304 | 0.009329 | -0.003838 | -0.004057 | 0.011732 | -0.012960 | -0.005264 | 0.024481 | -0.021813 | -0.020103 | 0.079306 | 0.056902 | -0.012589 |
| 2 | food_0 | disease_50 | 0.0 | 0 | 50 | 0.018382 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.1 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 41.508018 | -0.271734 | -0.049130 | -0.031626 | -0.067875 | -0.005537 | -0.014704 | -0.017518 | -0.034036 | -0.000631 | -0.004023 | 0.013805 | 0.000356 | -0.005335 | 0.003229 | 0.000713 | 0.000098 | -0.005014 | -0.005383 | -0.005519 | 0.003013 | -0.002217 | -0.009330 | -0.005702 | 0.001625 | -0.008246 | -0.001536 | -0.006982 | 0.013521 | 0.000661 | -0.003444 | 0.004891 | -0.000032 | -0.000943 | -0.001430 | 0.004226 | 0.000832 | 0.002662 | 0.003511 | 0.000865 | -0.001511 | 0.001598 | 0.003186 | -0.005701 | -0.000564 | 0.000999 | 0.000164 | 0.000142 | 0.002312 | 0.001523 | 0.002301 | -0.000096 | -0.000341 | 0.000267 | 0.001574 | -0.000061 | 0.000018 | -0.000039 | -0.000093 | -0.000128 | -0.000114 | -0.000045 | 0.000055 | -0.000003 | -8.250175e-06 | 0.000030 | 0.000038 | -0.000022 | 0.000025 | -0.000064 | -0.000052 | -0.000010 | 0.000063 | -0.000024 | 0.000188 | -0.000111 | -0.000098 | -0.000364 | -0.000028 | 0.000122 | -0.000164 | 0.000070 | -0.000014 | -0.000859 | -0.000266 | 0.000222 | 0.000684 | -0.000063 | -0.000192 | -0.000442 | 0.000511 | 0.000777 | 0.000287 | -0.000420 | 0.000969 | 0.001567 | 0.001200 | -0.000256 | 0.000491 | -0.000133 | -0.000430 | -0.001268 | -0.002388 | -0.000176 | -0.002063 | -0.002062 | 0.001144 | 0.001614 | 0.001952 | 0.002008 | 0.000060 | 0.001320 | -0.000544 | -0.000907 | 0.003609 | 0.001488 | -0.000267 | -0.002821 | 0.000381 | -0.003625 | 0.001080 | -0.003746 | -0.000740 | 0.004468 | 0.000185 | 0.005591 | -0.001899 | 0.001580 | 1.189615 | -0.109329 | 0.002665 | 0.190674 | 0.706729 | -0.526051 | 0.151012 | 1.535668 | 1.386908 | -0.136352 | 0.122367 | -0.631401 | 0.126397 | -0.276903 | 0.329921 | -0.058434 | 0.150027 | 0.081792 | -0.109353 | 0.098505 | -0.615293 | -0.061401 | 0.054187 | 0.171404 | 0.067380 | -0.178878 | 0.316629 | 0.142819 | -0.221195 | -0.169382 | -0.276687 | 0.346723 | -0.050159 | -0.171242 | -0.080506 | -0.118671 | 0.018256 | 0.047224 | -0.304223 | -0.152287 | 0.122938 | -0.058284 | -0.407862 | -0.010772 | -0.175505 | 0.132974 | 0.031009 | 0.221593 | -0.054744 | -0.009640 | -0.070451 | -0.264516 | 0.110601 | 0.291928 | 0.102618 | -0.166557 | -0.021385 | 0.064398 | 0.245133 | -0.163323 | -0.205366 | -0.078624 | -0.113051 | -0.086653 | 0.057236 | 0.089719 | 0.022555 | 0.033048 | 0.041558 | -0.154484 | -0.283011 | -0.139989 | -0.030849 | -0.023703 | -0.195976 | 0.047442 | 0.177886 | -0.031600 | -0.045078 | 0.172449 | -0.196692 | -0.226955 | -0.091421 | 0.116724 | 0.075902 | 0.198356 | 0.069463 | -0.098914 | -0.074465 | 0.018325 | 0.013663 | 0.044086 | 0.006720 | -0.092438 | 0.106865 | 0.042334 | -0.106831 | 0.047141 | -0.052669 | 0.032439 | -0.020543 | -0.067151 | 0.024235 | -0.023867 | -0.022965 | 0.079472 | -0.012705 | 0.022978 | 0.045083 | -0.016602 | 0.069385 | -0.034710 | -0.012632 | -0.055165 | -0.023180 | -0.105940 | -0.096701 | -0.139067 | 0.082889 | -0.008238 | -0.058866 | 0.072690 | -0.040983 | 0.001735 | -0.031011 | 0.012370 | -0.016576 | 0.062942 |
| 3 | food_0 | disease_1370 | 0.0 | 0 | 1370 | 0.214286 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.1 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 41.529790 | -0.260051 | -0.054846 | -0.039315 | -0.049941 | 0.002167 | -0.012543 | -0.030178 | -0.019744 | -0.009011 | -0.001326 | 0.012445 | 0.004044 | -0.010835 | -0.004046 | 0.019301 | 0.002939 | -0.006936 | -0.004579 | -0.001534 | -0.000085 | 0.007771 | -0.027475 | -0.001232 | 0.001875 | 0.000077 | -0.008815 | -0.020402 | -0.001688 | -0.008872 | -0.005646 | 0.016212 | 0.013411 | -0.018793 | 0.016397 | -0.003017 | -0.004065 | 0.003183 | -0.006690 | 0.018216 | -0.004084 | -0.009724 | 0.006243 | -0.004513 | -0.000231 | 0.000521 | -0.000797 | 0.003050 | -0.001297 | -0.004875 | -0.003577 | -0.001533 | -0.001472 | 0.001390 | 0.000372 | -0.000034 | -0.000062 | -0.000605 | 0.000263 | -0.000107 | 0.000003 | 0.000030 | 0.000036 | 0.000024 | -8.291036e-07 | -0.000040 | -0.000080 | 0.000041 | -0.000044 | -0.000096 | -0.000057 | 0.000017 | 0.000058 | -0.000022 | -0.000132 | 0.000107 | 0.000208 | 0.000220 | -0.000585 | -0.000699 | -0.000664 | 0.000296 | -0.000517 | 0.000028 | 0.000650 | -0.000914 | -0.000190 | 0.000124 | -0.001585 | -0.000599 | -0.000961 | 0.000682 | 0.000873 | -0.000953 | 0.001415 | -0.004218 | 0.004151 | 0.003551 | 0.000414 | -0.001865 | -0.000427 | -0.000561 | 0.000712 | 0.000712 | -0.002766 | -0.002378 | 0.000731 | -0.000705 | 0.004565 | -0.001887 | -0.005995 | 0.003522 | 0.002315 | -0.002299 | 0.004152 | -0.007743 | 0.002656 | -0.005594 | -0.001302 | 0.003646 | -0.001710 | 0.006324 | 0.006646 | -0.006136 | 0.003705 | 0.010991 | -0.006933 | 0.001896 | 1.070765 | -0.808722 | -0.295265 | -0.377572 | 0.224404 | 0.624164 | -0.171772 | -0.033262 | -0.001275 | -0.288270 | -0.386495 | 0.179418 | 0.970317 | -0.800248 | -1.032255 | 0.141866 | -0.130448 | 0.211681 | -0.161183 | -0.133450 | 0.103317 | -0.330761 | -0.286104 | -0.109326 | 0.120395 | 0.188776 | 0.892057 | -0.053633 | 0.402967 | 0.386931 | -0.284458 | -0.034337 | 0.094869 | -0.405306 | -0.019650 | 0.119014 | -0.220812 | -0.157350 | 0.113438 | 0.337078 | -0.105879 | -0.191973 | 0.182164 | 0.150012 | -0.002031 | 0.214086 | -0.001215 | 0.217673 | -0.046450 | -0.045357 | 0.213626 | -0.012232 | 0.095100 | 0.030139 | 0.068430 | -0.192951 | -0.114827 | 0.035296 | -0.216153 | 0.158659 | 0.111714 | -0.154277 | -0.135024 | 0.234255 | -0.090858 | 0.091808 | 0.030877 | -0.000725 | -0.186567 | -0.036827 | -0.024478 | 0.017350 | -0.110065 | -0.012707 | -0.191022 | 0.007126 | -0.027630 | 0.049433 | 0.232110 | -0.107700 | 0.023428 | -0.015442 | -0.073996 | -0.168445 | -0.069636 | 0.062691 | 0.074576 | 0.089530 | -0.082161 | -0.074518 | -0.042437 | 0.031088 | -0.155015 | 0.019656 | 0.039321 | -0.055407 | -0.063896 | -0.093685 | 0.226617 | 0.113530 | -0.045031 | -0.174211 | -0.128222 | -0.114841 | -0.031775 | 0.003285 | -0.002169 | -0.108374 | 0.002132 | 0.015701 | -0.025789 | 0.049046 | -0.074700 | 0.020771 | -0.000206 | 0.066936 | 0.028114 | -0.094354 | -0.114083 | 0.041070 | -0.060834 | 0.031545 | 0.020605 | 0.045850 | 0.060787 | -0.054691 | -0.081137 | -0.041880 |
| 4 | food_0 | disease_1015 | 0.0 | 0 | 1015 | 0.202749 | 8.2 | 0.157 | 0.92 | 27.0 | 0.481 | 23.0 | 79.0 | 6.0 | 0.076 | 0.126 | 2.1 | 3.99 | 70.0 | 32.0 | 0.96 | 92.82 | 0.69 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 1.194324 | 0.016060 | 0.021213 | -0.019584 | 0.053625 | 0.013038 | 0.009994 | -0.015606 | 0.062566 | -0.061401 | 0.022027 | -0.038624 | -0.055178 | -0.048019 | 0.759425 | 0.398878 | 0.111763 | 0.015837 | 0.054869 | -0.047739 | 0.036110 | -0.022543 | 0.029236 | -0.030270 | -0.006784 | -0.003725 | -0.032971 | 0.016045 | 0.015254 | -0.008581 | -0.012702 | -0.011345 | -0.010733 | 0.009206 | -0.009229 | 0.000618 | 0.005975 | -0.003449 | -0.007818 | -0.009403 | 0.007591 | 0.002266 | -0.004004 | -0.002725 | -0.003839 | 0.002820 | 0.000835 | -0.002778 | 0.000629 | -0.002743 | -0.000550 | 0.000052 | 0.000554 | -0.001329 | 0.000971 | 0.000060 | 0.000091 | 0.000130 | -0.000430 | -0.000074 | -0.000154 | -0.000046 | 0.000024 | 0.000015 | -4.640532e-06 | -0.000004 | 0.000006 | 0.000035 | 0.000012 | -0.000102 | -0.000041 | -0.000042 | 0.000003 | 0.000004 | 0.000252 | -0.000294 | -0.000062 | -0.000560 | 0.000023 | -0.000038 | -0.000145 | 0.000341 | 0.000053 | -0.001102 | 0.000195 | -0.000488 | -0.000224 | 0.000328 | -0.000370 | 0.000285 | 0.001096 | -0.000158 | 0.001338 | 0.000476 | 0.002296 | 0.000573 | 0.001374 | -0.001213 | -0.000561 | 0.000960 | -0.000290 | -0.000561 | -0.001062 | -0.001659 | 0.000637 | -0.004378 | -0.002726 | 0.002012 | 0.000543 | 0.001945 | 0.005084 | 0.001443 | 0.000259 | -0.000784 | 0.001073 | -0.002402 | 0.005187 | 0.002465 | -0.000312 | -0.000665 | 0.002359 | 0.003525 | 0.000040 | 0.003171 | -0.006081 | -0.004884 | 0.008354 | -0.002090 | 1.286808 | -0.267558 | 0.293177 | 0.179919 | 0.753527 | -0.548435 | -0.262182 | 0.455970 | 0.503655 | -0.740368 | -0.459004 | -0.473660 | 0.816256 | -0.538978 | 1.083409 | 0.272414 | -0.283723 | -0.921948 | 0.089385 | -0.305596 | -0.141462 | 0.209232 | -0.102136 | 0.361745 | -0.073097 | 0.247010 | -0.215279 | 0.622676 | -0.036221 | -0.100733 | -0.095383 | 0.252611 | 0.112919 | -0.032485 | -0.269291 | 0.179181 | -0.179328 | -0.041581 | -0.208619 | -0.143577 | -0.258412 | -0.121063 | -0.410299 | 0.081461 | -0.118664 | 0.432868 | 0.512245 | -0.090577 | 0.357983 | -0.083231 | 0.062759 | 0.106986 | 0.043856 | -0.294729 | -0.021793 | 0.070910 | -0.007633 | -0.070904 | 0.097167 | 0.162714 | 0.052339 | 0.102275 | 0.007705 | -0.093869 | -0.008447 | 0.046357 | 0.154401 | -0.124018 | -0.049133 | 0.308927 | -0.101389 | 0.107464 | 0.126912 | -0.087374 | 0.120835 | -0.130838 | -0.116480 | -0.070292 | -0.049393 | 0.028696 | 0.104959 | 0.181812 | 0.097010 | -0.075481 | 0.084535 | -0.122547 | 0.046716 | 0.049885 | -0.094828 | 0.062870 | -0.062986 | 0.030545 | 0.001237 | -0.047724 | -0.022991 | 0.004514 | 0.055843 | 0.048817 | 0.044805 | 0.058769 | -0.070773 | 0.088568 | 0.020527 | -0.016702 | 0.049144 | -0.057848 | -0.151351 | -0.153255 | 0.007800 | 0.052151 | 0.053366 | 0.039268 | -0.006772 | 0.077348 | -0.028753 | -0.118866 | 0.028853 | -0.078241 | -0.035130 | -0.033633 | -0.015799 | -0.042921 | 0.035031 | 0.071032 | -0.097231 | 0.081445 | -0.014732 | 0.010713 |
- 1
3.2.3 交叉特征
这里按特征重要性选取靠前的部分特征进行交叉
topn = ['N_33', 'N_198', 'N_74','N_188','N_82','N_42','N_111','disease','food']
for i in range(len(topn)):
for j in range(i + 1, len(topn)):
data[f'{topn[i]}+{topn[j]}'] = data[topn[i]] + data[topn[j]]
data[f'{topn[i]}-{topn[j]}'] = data[topn[i]] - data[topn[j]]
data[f'{topn[i]}*{topn[j]}'] = data[topn[i]] * data[topn[j]]
data[f'{topn[i]}/{topn[j]}'] = data[topn[i]] / (data[topn[j]]+1e-5)
- 1
- 2
- 3
- 4
- 5
- 6
- 7
drop_cols = ['disease_id', 'food_id', 'related']
- 1
3.2.3 特征筛选
去除掉只有单一取值的特征
for f in data.columns:
if data[f].nunique() < 2:
drop_cols.append(f)
- 1
- 2
- 3
test_df = data[data["related"].isnull() == True].copy().reset_index(drop=True)
train_df = data[~data["related"].isnull() == True].copy().reset_index(drop=True)
- 1
- 2
feature_name = [f for f in train_df.columns if f not in drop_cols]
X_train = train_df[feature_name].reset_index(drop=True)
X_test = test_df[feature_name].reset_index(drop=True)
y = train_df['related'].reset_index(drop=True)
print(len(feature_name))
print(feature_name)
- 1
- 2
- 3
- 4
- 5
- 6
548
['food', 'disease', 'disease_related_mean', 'N_198', 'N_33', 'N_211', 'N_82', 'N_101', 'N_42', 'N_111', 'N_165', 'N_177', 'N_146', 'N_17', 'N_113', 'N_106', 'N_14', 'N_74', 'N_209', 'N_188', 'disease1_svd_0', 'disease1_svd_1', 'disease1_svd_2', 'disease1_svd_3', 'disease1_svd_4', 'disease1_svd_5', 'disease1_svd_6', 'disease1_svd_7', 'disease1_svd_8', 'disease1_svd_9', 'disease1_svd_10', 'disease1_svd_11', 'disease1_svd_12', 'disease1_svd_13', 'disease1_svd_14', 'disease1_svd_15', 'disease1_svd_16', 'disease1_svd_17', 'disease1_svd_18', 'disease1_svd_19', 'disease1_svd_20', 'disease1_svd_21', 'disease1_svd_22', 'disease1_svd_23', 'disease1_svd_24', 'disease1_svd_25', 'disease1_svd_26', 'disease1_svd_27', 'disease1_svd_28', 'disease1_svd_29', 'disease1_svd_30', 'disease1_svd_31', 'disease1_svd_32', 'disease1_svd_33', 'disease1_svd_34', 'disease1_svd_35', 'disease1_svd_36', 'disease1_svd_37', 'disease1_svd_38', 'disease1_svd_39', 'disease1_svd_40', 'disease1_svd_41', 'disease1_svd_42', 'd
- 1
- 2
print(test_df.shape)
- 1
(47212, 551)
- 1
3.2.3 模型训练
本次仅使用lightgbm模型来训练。
train_pred = {}
test_pred = {}
- 1
- 2
seeds = [2] num_model_seed = 1 oof = np.zeros(X_train.shape[0]) prediction = np.zeros(X_test.shape[0]) feat_imp_df = pd.DataFrame({'feats': feature_name, 'imp': 0}) parameters = { 'learning_rate': 0.05, 'boosting_type': 'gbdt', 'objective': 'binary', 'metric': 'auc', 'num_leaves': 63, 'feature_fraction': 0.8, 'bagging_fraction': 0.8, 'bagging_freq': 5, 'seed': 2022, 'bagging_seed': 1, 'feature_fraction_seed': 7, 'min_data_in_leaf': 20, 'verbose': -1, 'n_jobs':8, } fold = 5 for model_seed in range(num_model_seed): print(seeds[model_seed],"--------------------------------------------------------------------------------------------") oof_cat = np.zeros(X_train.shape[0]) prediction_cat = np.zeros(X_test.shape[0]) skf = StratifiedKFold(n_splits=fold, random_state=seeds[model_seed], shuffle=True) for index, (train_index, test_index) in enumerate(skf.split(X_train, y)): train_x, test_x, train_y, test_y = X_train[feature_name].iloc[train_index], X_train[feature_name].iloc[test_index], y.iloc[train_index], y.iloc[test_index] dtrain = lgb.Dataset(train_x, label=train_y) dval = lgb.Dataset(test_x, label=test_y) lgb_model = lgb.train( parameters, dtrain, num_boost_round=10000, valid_sets=[dval], early_stopping_rounds=100, verbose_eval=100, ) oof_cat[test_index] += lgb_model.predict(test_x,num_iteration=lgb_model.best_iteration) prediction_cat += lgb_model.predict(X_test,num_iteration=lgb_model.best_iteration) / fold feat_imp_df['imp'] += lgb_model.feature_importance() del train_x del test_x del train_y del test_y del lgb_model oof += oof_cat / num_model_seed prediction += prediction_cat / num_model_seed gc.collect()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
- 42
- 43
- 44
- 45
- 46
- 47
- 48
- 49
- 50
- 51
- 52
3.2.4 结果可视化
train_pred['lgb'] = oof
test_pred['lgb'] = prediction
- 1
- 2
print("lgb train auc: ", roc_auc_score(y, train_pred['lgb']))
- 1
lgb train auc: 0.9778226537246766
- 1
scores = []; thresholds = []
best_score = 0; best_threshold = 0
for threshold in np.arange(0.1,0.9,0.01):
print(f'{threshold:.02f}, ',end='')
preds = (train_pred['lgb'].reshape((-1)) > threshold).astype('int')
m = f1_score(y.values.reshape((-1)), preds, average='binary')
scores.append(m)
thresholds.append(threshold)
if m>best_score:
best_score = m
best_threshold = threshold
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
0.10, 0.11, 0.12, 0.13, 0.14, 0.15, 0.16, 0.17, 0.18, 0.19, 0.20, 0.21, 0.22, 0.23, 0.24, 0.25, 0.26, 0.27, 0.28, 0.29, 0.30, 0.31, 0.32, 0.33, 0.34, 0.35, 0.36, 0.37, 0.38, 0.39, 0.40, 0.41, 0.42, 0.43, 0.44, 0.45, 0.46, 0.47, 0.48, 0.49, 0.50, 0.51, 0.52, 0.53, 0.54, 0.55, 0.56, 0.57, 0.58, 0.59, 0.60, 0.61, 0.62, 0.63, 0.64, 0.65, 0.66, 0.67, 0.68, 0.69, 0.70, 0.71, 0.72, 0.73, 0.74, 0.75, 0.76, 0.77, 0.78, 0.79, 0.80, 0.81, 0.82, 0.83, 0.84, 0.85, 0.86, 0.87, 0.88, 0.89,
- 1
import matplotlib.pyplot as plt
# PLOT THRESHOLD VS. F1_SCORE
plt.figure(figsize=(20,5))
plt.plot(thresholds,scores,'-o',color='blue')
plt.scatter([best_threshold], [best_score], color='blue', s=300, alpha=1)
plt.xlabel('Threshold',size=14)
plt.ylabel('Validation F1 Score',size=14)
plt.title(f'Threshold vs. F1_Score with Best F1_Score = {best_score:.3f} at Best Threshold = {best_threshold:.3}',size=18)
plt.show()
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-JhOW2ktx-1682508721966)(main_files/main_77_0.png)]
auc = roc_auc_score(y, train_pred['lgb'])
f1 = best_score
print((auc + f1) / 2)
- 1
- 2
- 3
0.8939347040505519
- 1
`
0.8939347040505519
- 1
3.2.5 生成提交结果
控制1的个数为4100个左右,最终结果:
# label=[1 if x >= 0.265+0.235 else 0 for x in prediction+0.235]
# np.sum(label)
label=[1 if x >= 0.26+0.24 else 0 for x in prediction+0.25]
np.sum(label)
- 1
- 2
- 3
- 4
- 5
4032
- 1
preliminary_a_submit_sample['related_prob'] = prediction+0.25
- 1
preliminary_a_submit_sample.to_csv('submit.csv', index=False)
- 1
四、项目总结
4.1 提交结果

4.2 优化思路
-
由于foodid只有训练集有,那么是否可以使用food侧的特征,做相似度模型,例如共现矩阵、tfidf、embedding等。
-
目标编码做了之后,线下会涨很多但是线上长得比较少,还是过拟合比较严重,是否可以考虑根据疾病特征做聚类,然后减轻这种情况。同理food侧特征也可以做聚类,用来解决测试集都是训练集未曾出现过的id的问题。
-
交叉特征里面,对于food侧只取了一部分,是否可以多取一点(进一步上述base可以通过筛选food特征提升至7976的分数,但是会很抖)。
-
特征筛选的地方,并没有剔除缺失率高的特征,也没有根据对抗验证进行筛选,或许可以进行尝试。
-
模型训练的参数,学习率太低,叶子节点数太高,导致模型过拟合比较严重,可以考虑调参(可以上分)。
-
目前仅使用了lightgbm模型,可以考虑xgboost,catboost模型,进行模型的集成(xgb貌似还不错,需要祖传参数)。
-
上述base是1的个数为4100个,可以调整不同的个数来测试,亲测不同的特征组合哪怕只是添加一个,最优的1的个数都是不同的。
-
svd的维度是可以调整的,不一定每个都是一样的。以及还可以使用pca的方法,不过需要进行归一化。
4.3 作者介绍
本人是AI达人特训营第三期项目中的一名学员,非常有幸能与大家分享自己的所思所想。
作者:范远展 指导导师:黄灿桦
此文章为搬运
原项目链接



