- 1vue+elementui实现导航栏三级菜单动态显示_三级动态
- 2保研的完整流程你清楚吗?_保研流程及详细攻略
- 3华为OD机试C、D卷 - 德州扑克(Java & JS & Python & C & C++)_error: could not find a version that satisfies the
- 4SpringBoot:整合Swagger3.0与RESTful接口整合返回值(2020最新最易懂)_swagger3.0版本返回结果带名字
- 5spring boot+vue3整合项目(后端)_springboot vue3
- 6解决Yolov5-7-0 运行classify/train.py时,报错:AttributeError: module ‘PIL.Image‘ has no attribute ‘ANTIALIAS‘_yolo module 'pil.image' has no attribute 'antialia
- 7记录解决select into outfile&load data报错ERROR 1(Errcode: 13 - Permission denied)
- 8总结 UDP 的报文结构和注意事项
- 9oracle_Listener refused the connection_centos oracle listener refused the connection with
- 10.NET 4.0运行.NET 2.0兼容方法 (2)
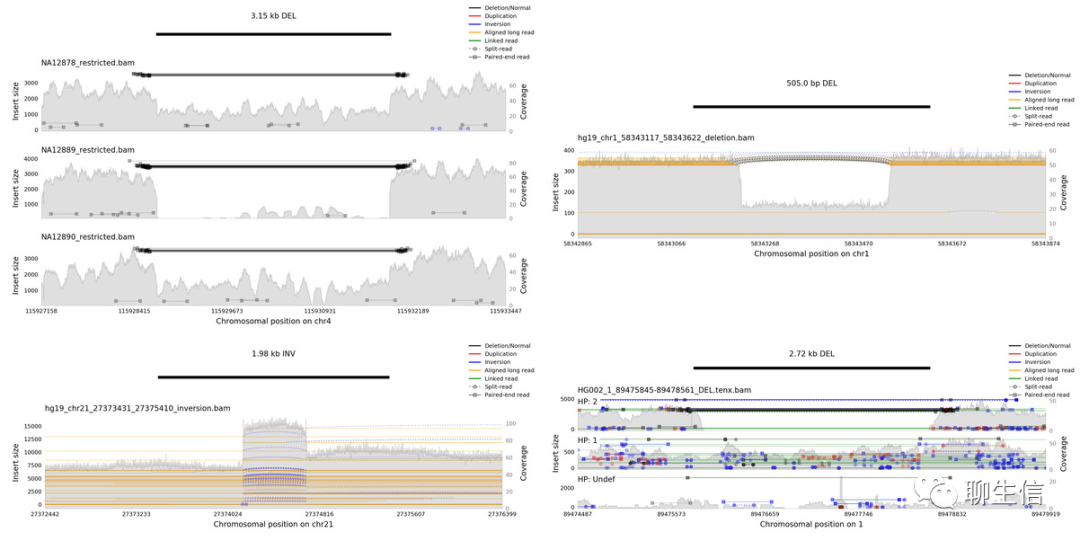
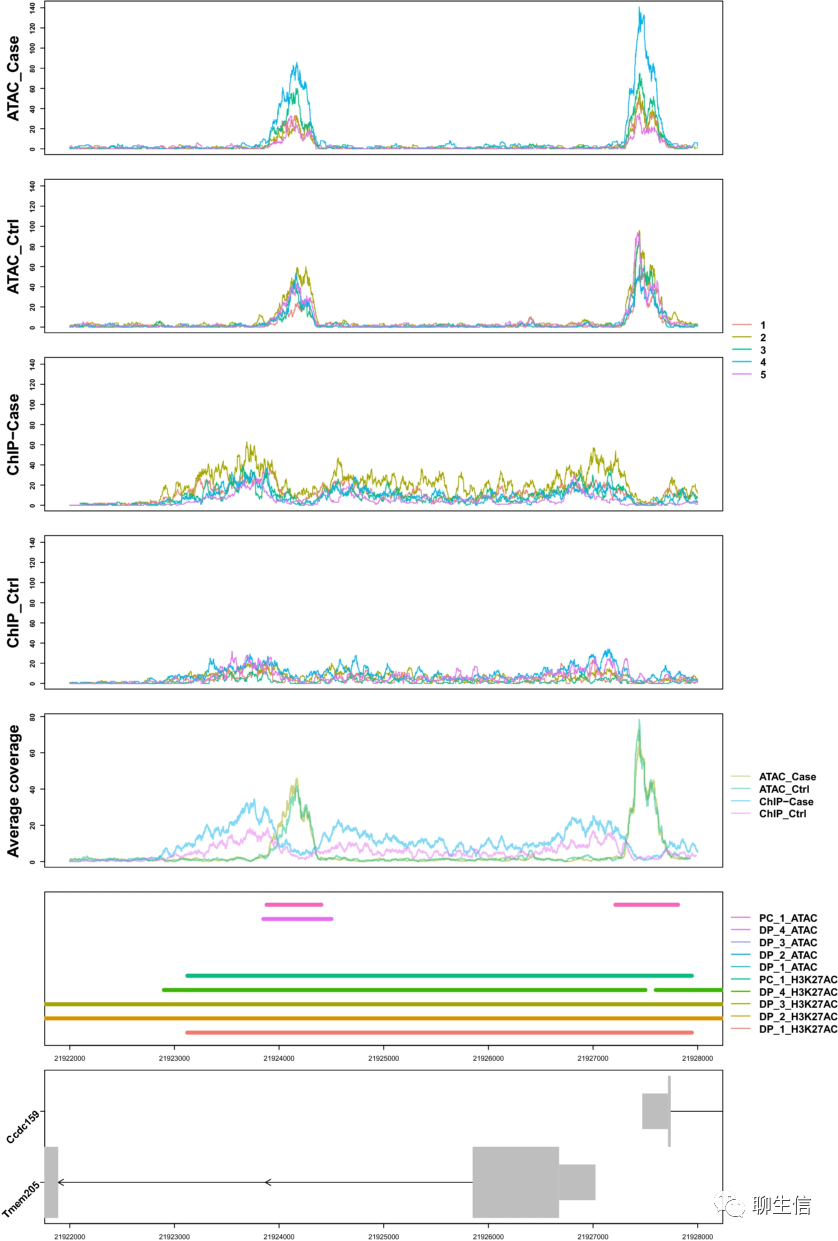
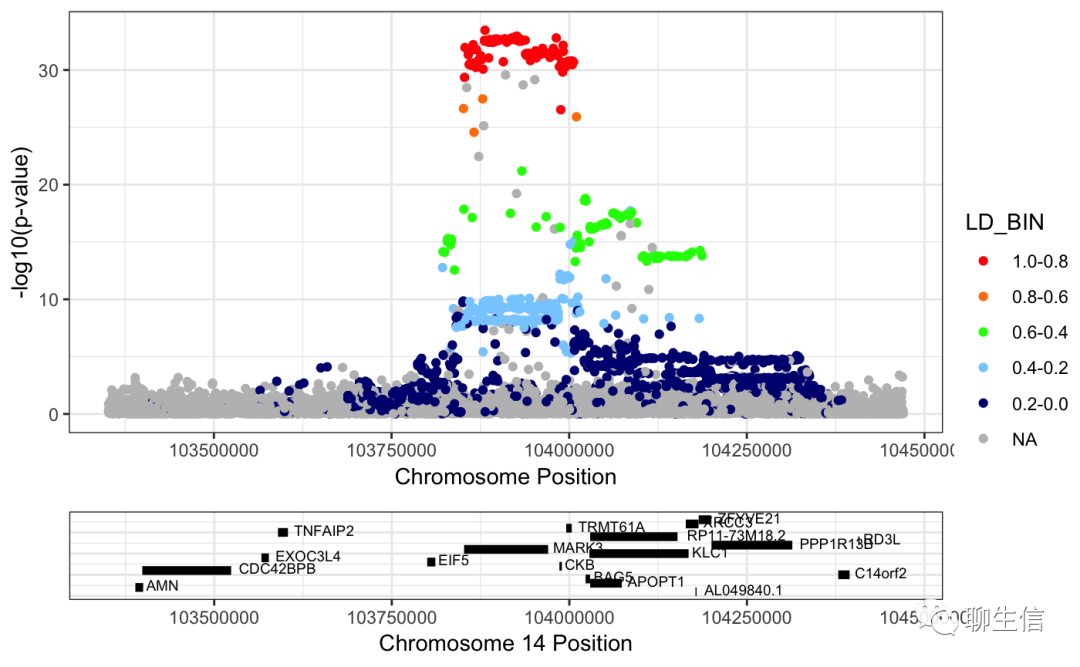
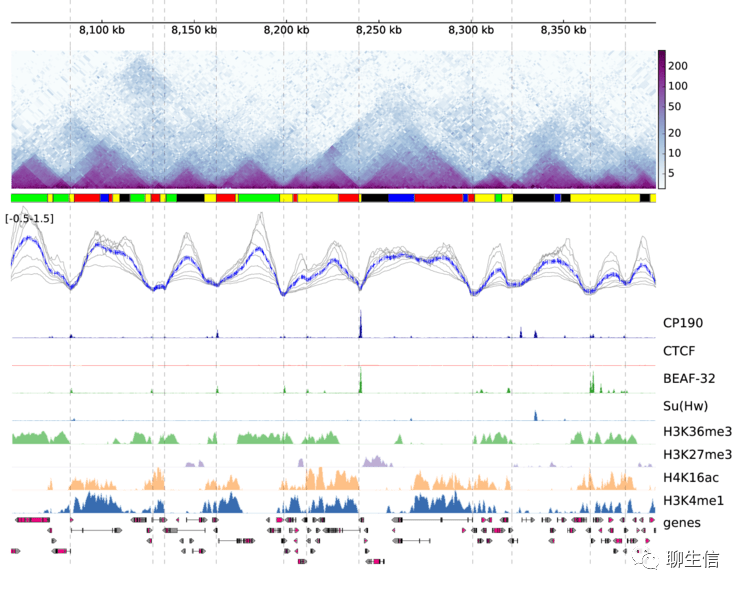
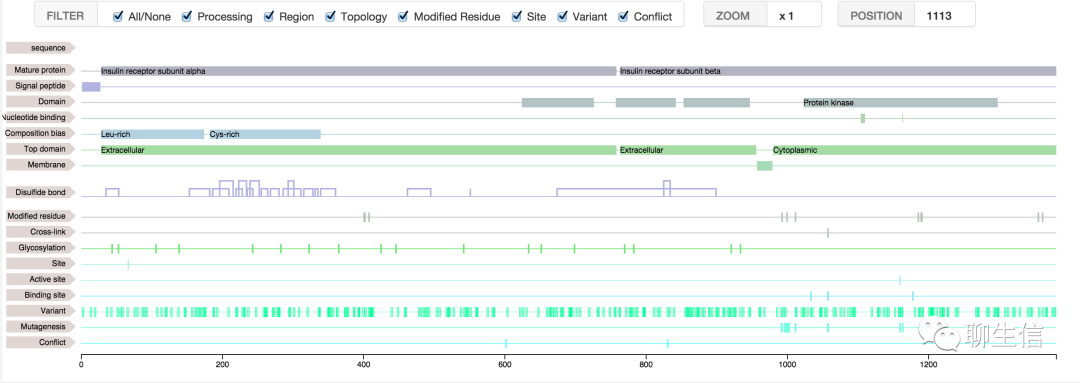
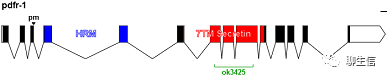
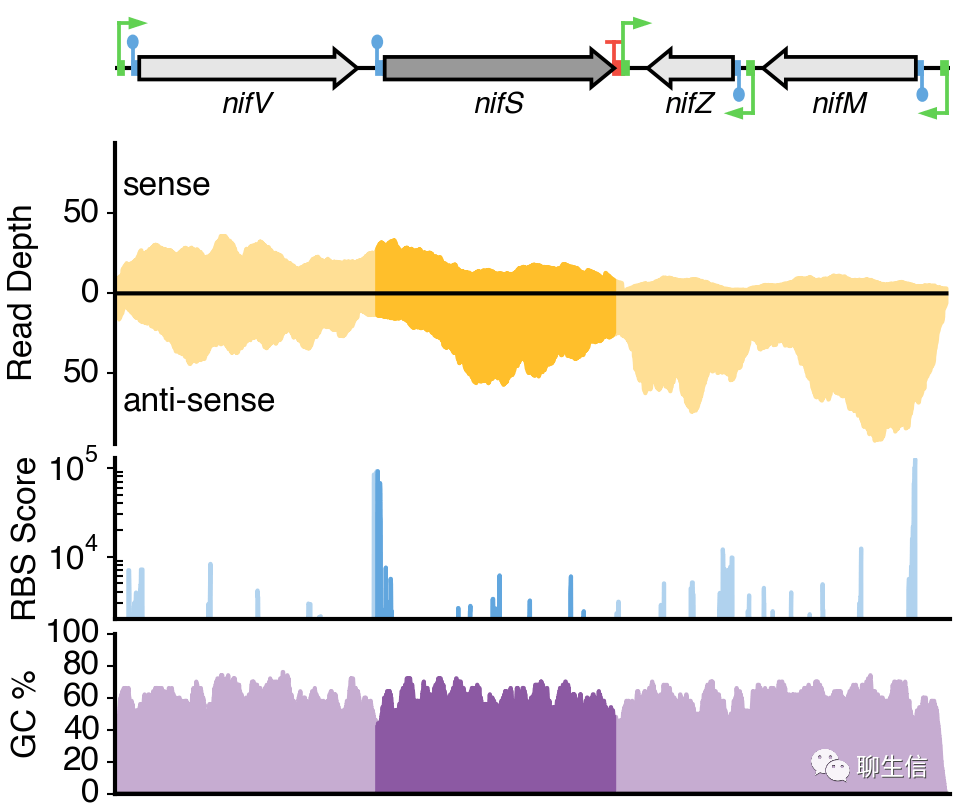
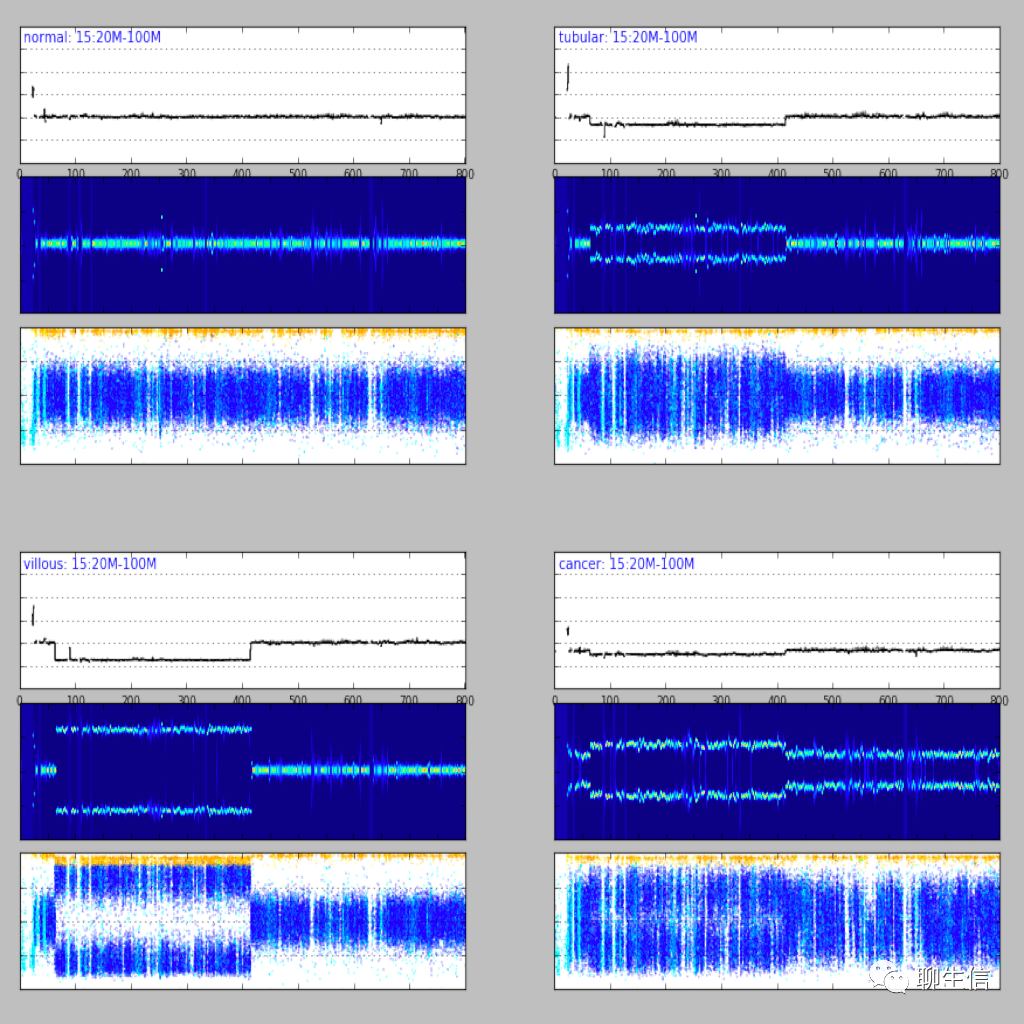
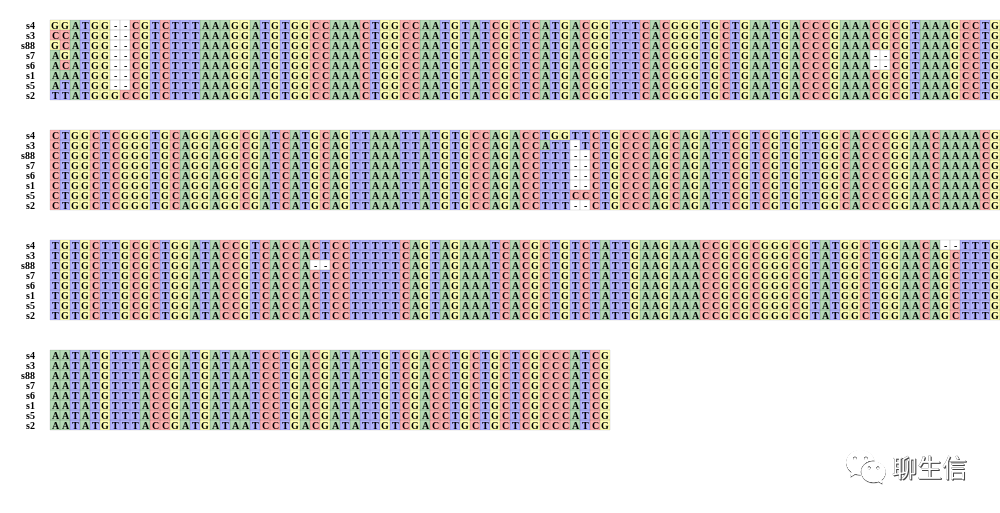
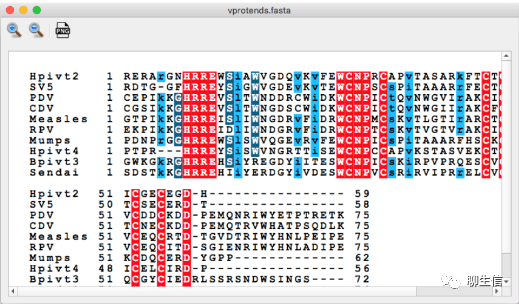
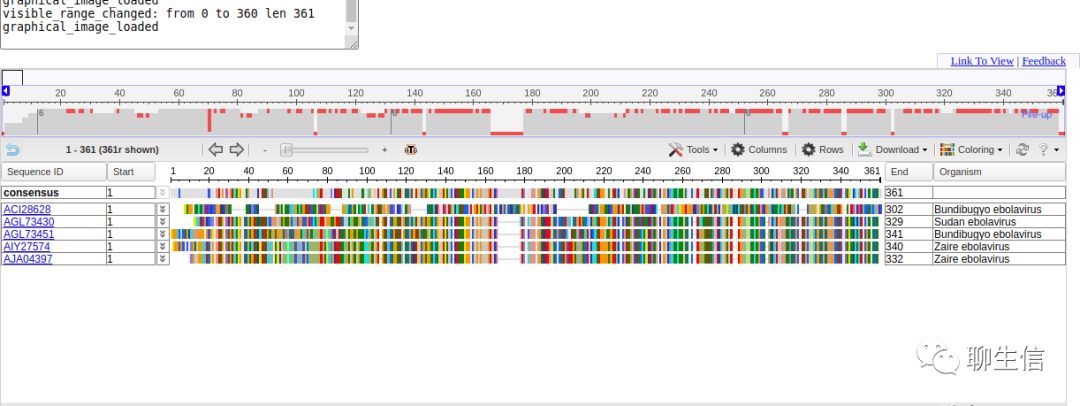
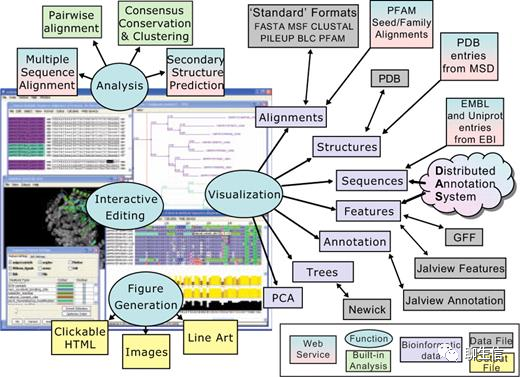
346个基因组可视化工具一网打进!_flexidot
赞
踩
Github来源:https://github.com/cmdcolin/awesome-genome-visualization
配套网页:
https://cmdcolin.github.io/awesome-genome-visualization/?latest=true
工具名称、下载链接、文献、编程语言、运行平台如下
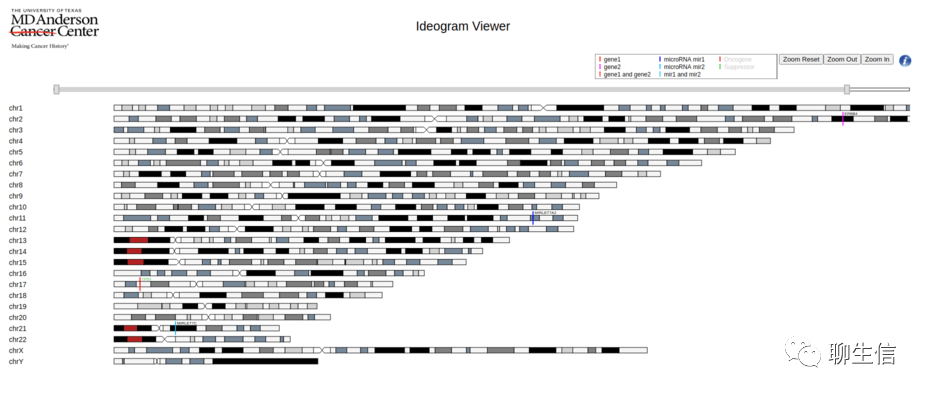
Ideogram viewer
https://bioinformatics.mdanderson.org/public-software/ideogramviewer/
Language: JS
Platform: Web
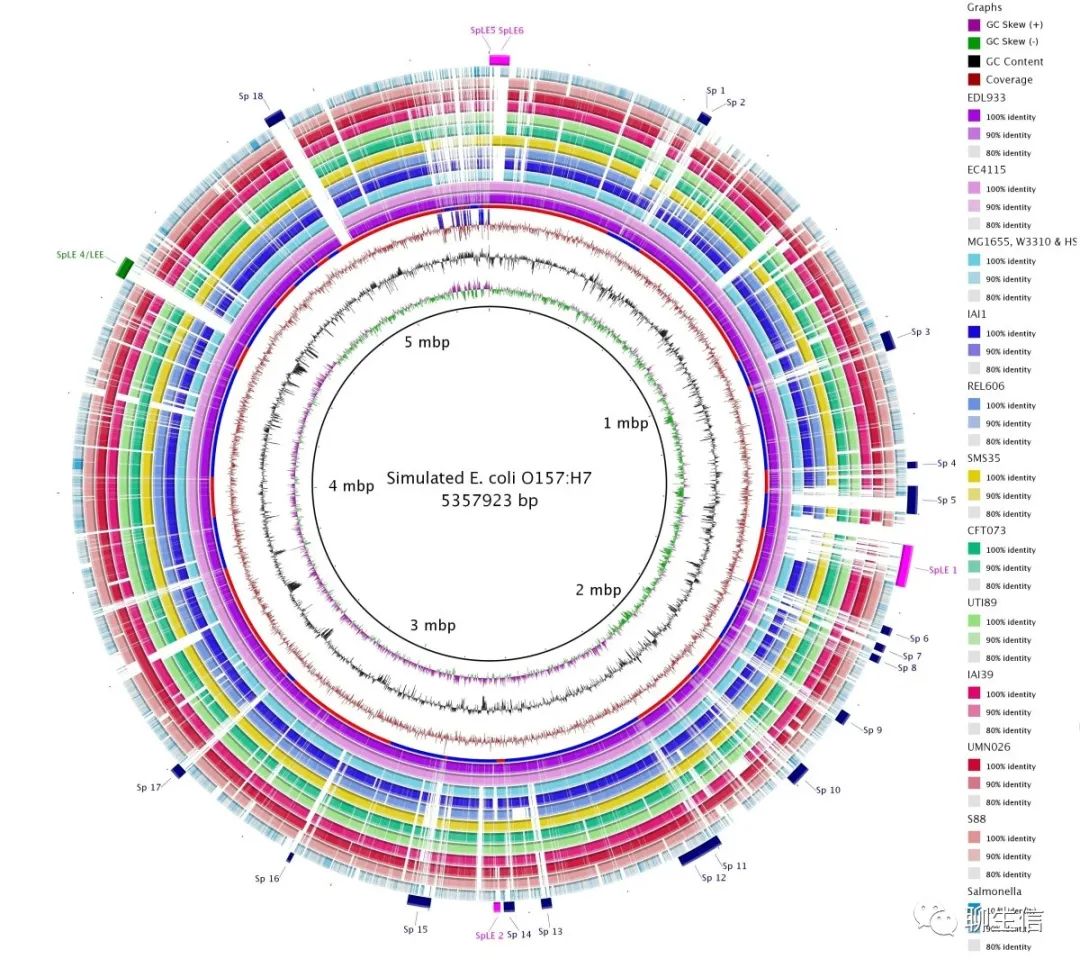
BRIG
http://brig.sourceforge.net
Publication: https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-12-402
Language: Java
Tags: Comparative, Circular
Platform: Desktop
GenomeMaps
http://www.genomemaps.org/
Publication: https://academic.oup.com/nar/article/41/W1/W41/1113984
Language: JS
Tags: General, Deadlink

gcMapExplorer
https://github.com/rjdkmr/gcMapExplorer
Language: Python
Tags: Hi-C
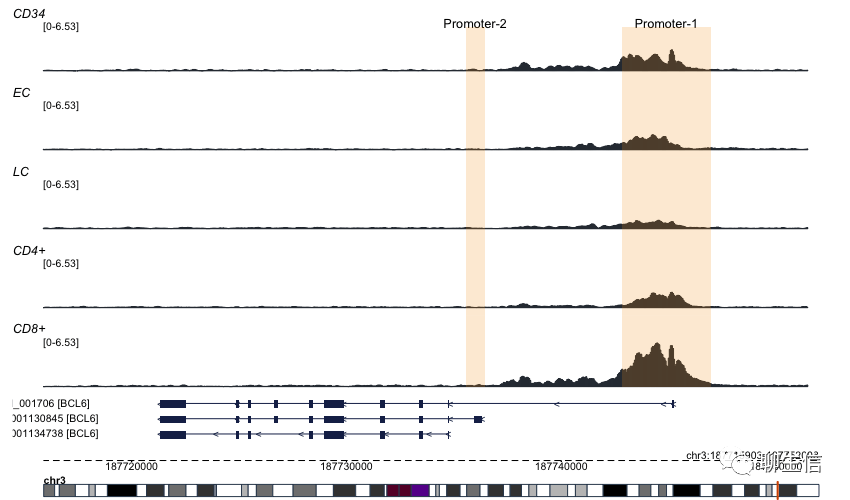
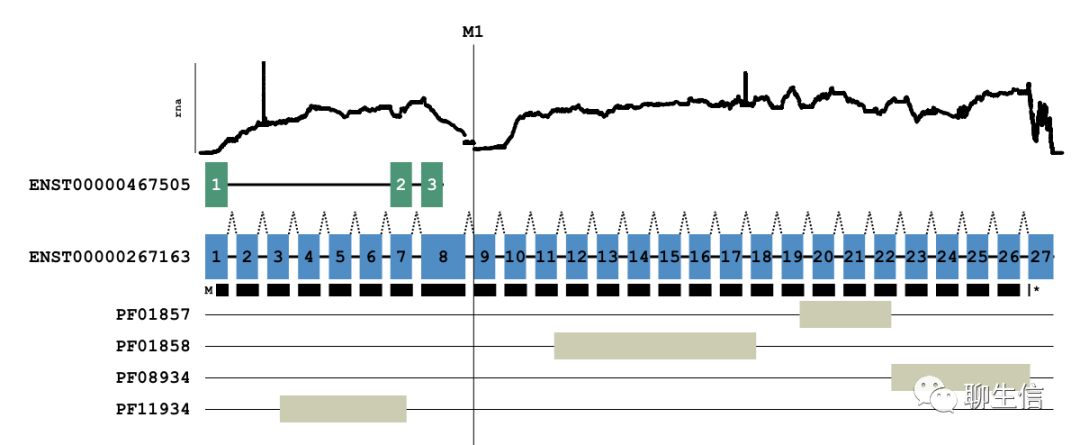
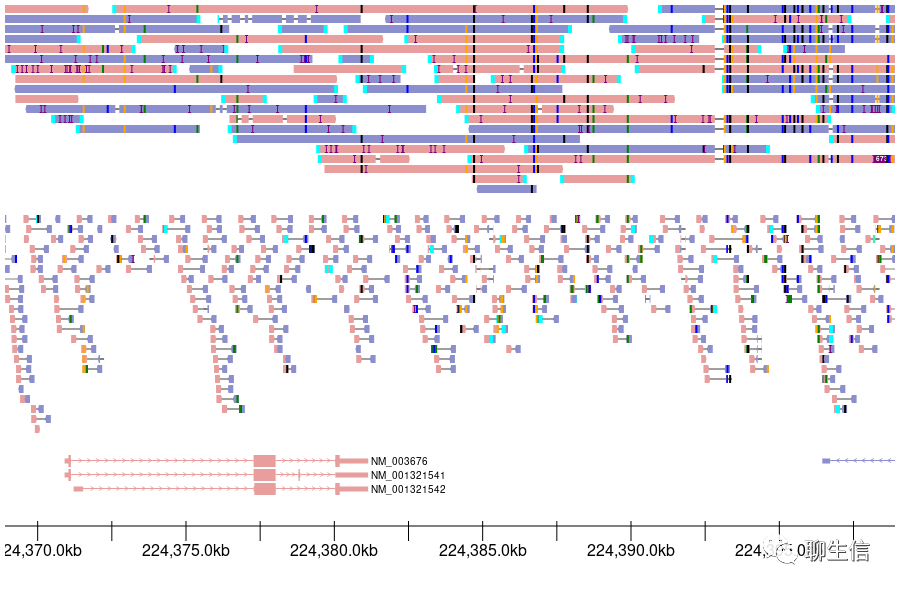
Trackplot
https://github.com/PoisonAlien/trackplot
Language: R
Tags: Coverage, Gene structure
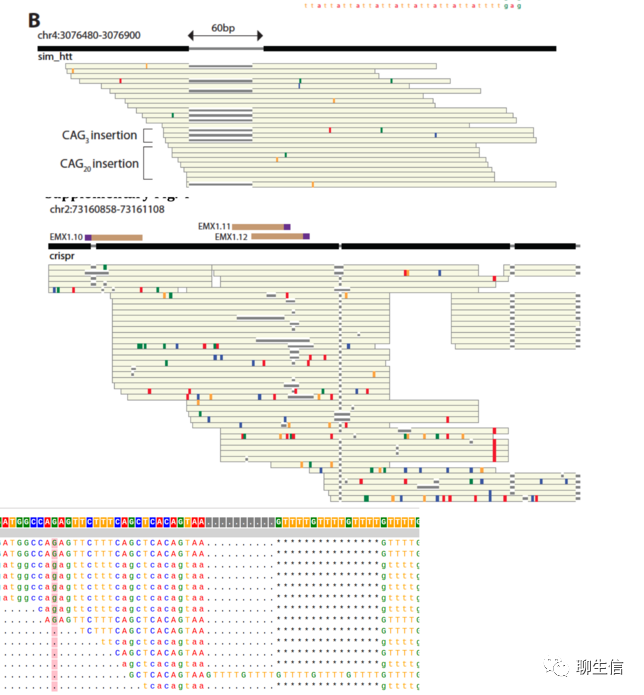
REViewer
https://github.com/Illumina/REViewer
Publication: https://www.biorxiv.org/content/10.1101/2021.10.20.465046v1.full.pdf
Tags: Repeats
Note: See also GraphAlignmentViewer, similar look and from illumina also
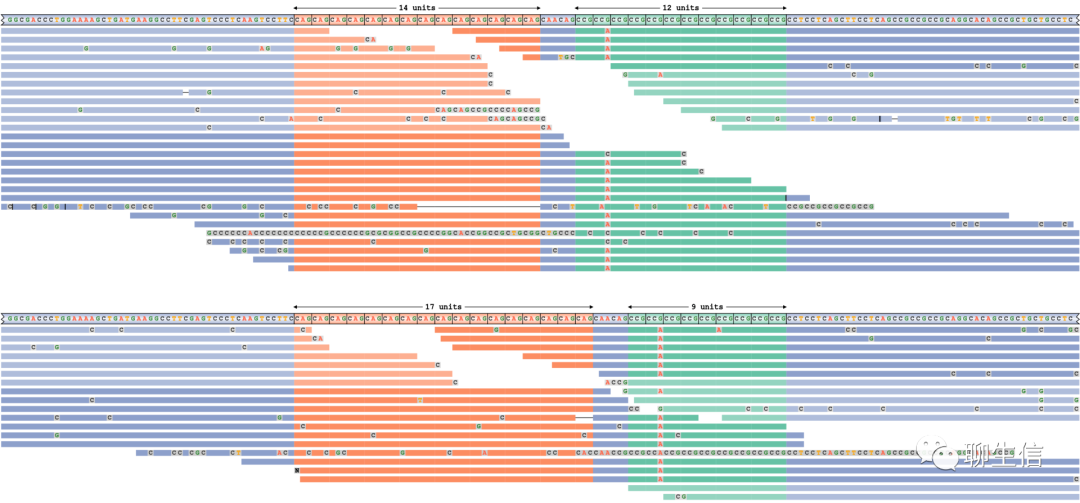
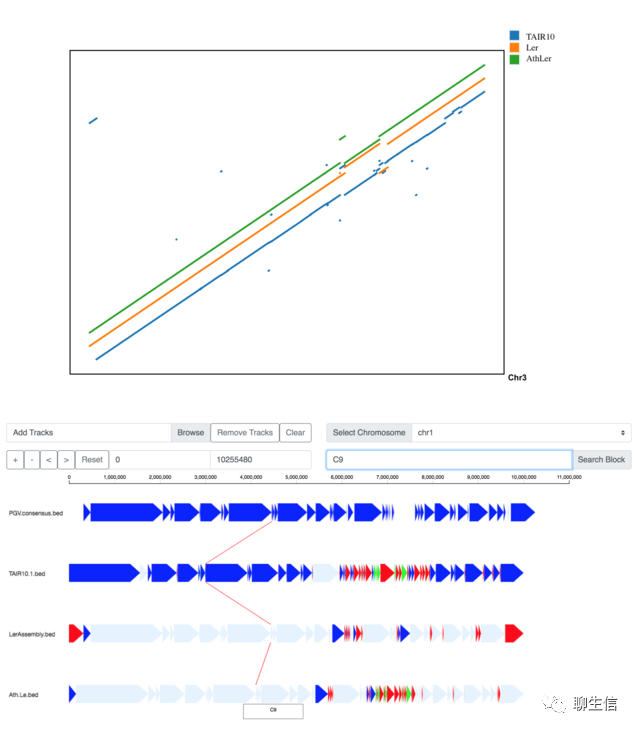
PGV
https://github.com/ucrbioinfo/PGV
Publication: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-021-04424-w
Language: Python
Tags: Pangenome, Dotplot, Comparative
Platform: Web
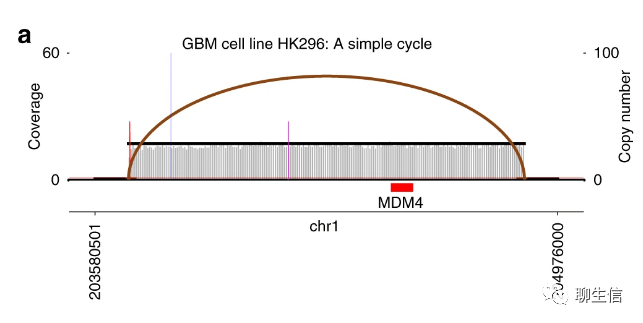
AmpliconArchitect
https://github.com/virajbdeshpande/AmpliconArchitect
Language: Python
Tags: SV, Cancer
Note: The term amplicon refers essentially to extrachromosomal DNA in cancer
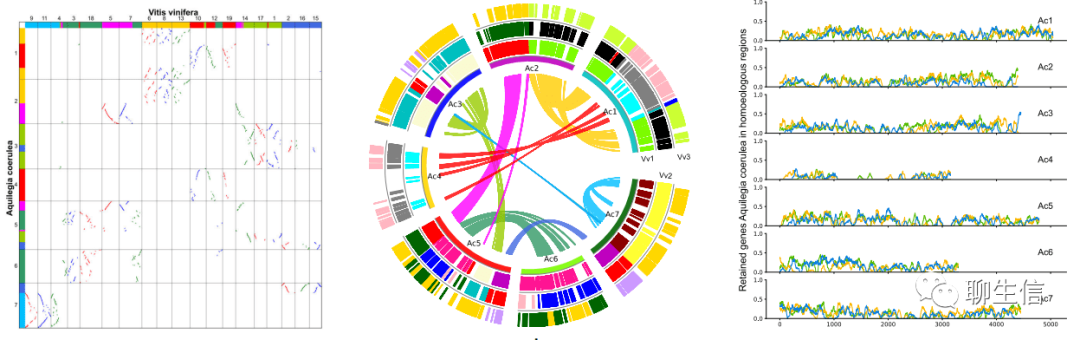
WGDI
https://github.com/SunPengChuan/wgdi
Publication: https://www.biorxiv.org/content/10.1101/2021.04.29.441969v1.full.pdf
Language: Python
Tags: Comparative, Dotplot, Circular
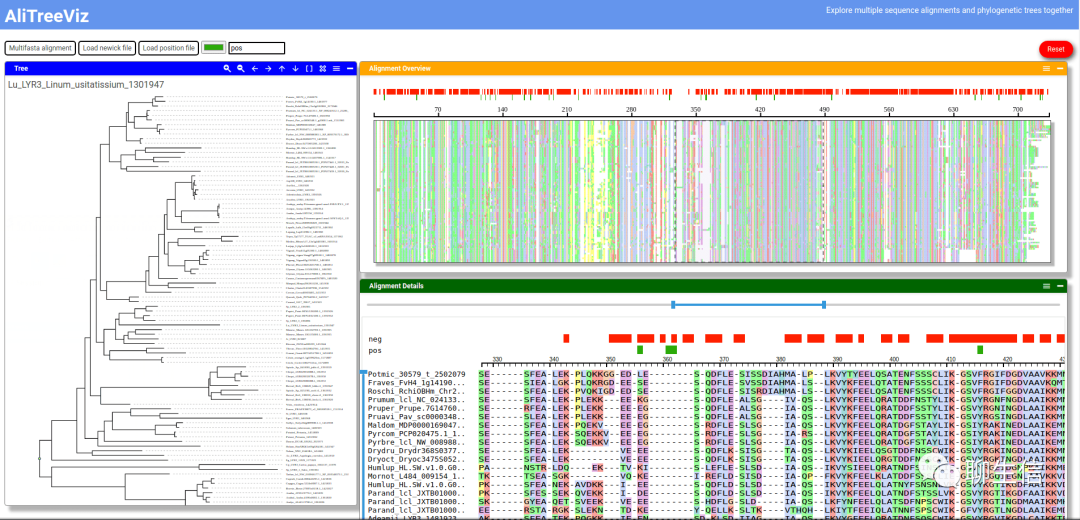
AliTreeViz
https://www.npmjs.com/package/alitreeviz
Language: JS
Tags: MSA, Phylogenetics
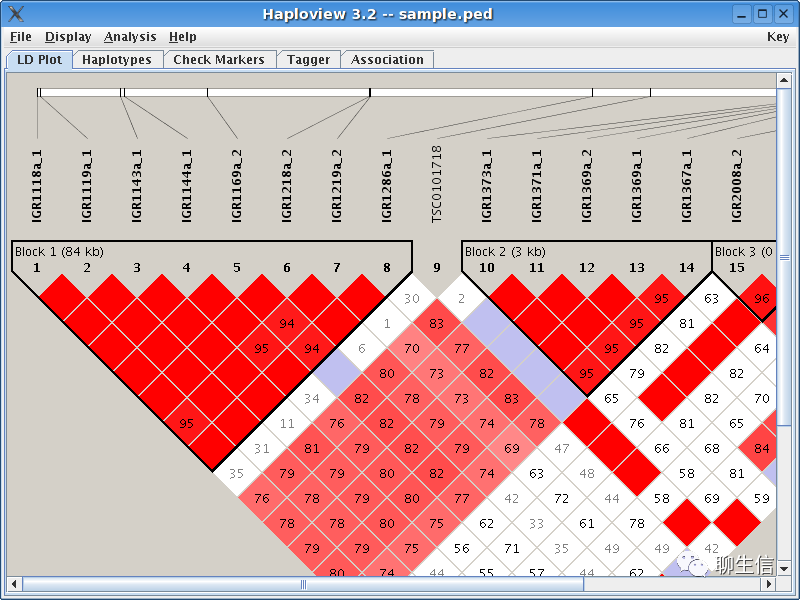
Haploview
https://www.broadinstitute.org/haploview/haploview
Language: Java
Platform: Desktop
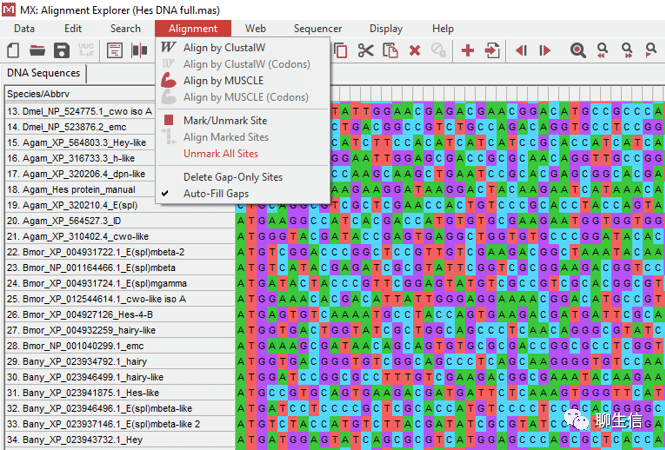
MEGA-X
https://www.megasoftware.net/
Tags: MSA, Phylogenetics
Platform: Desktop
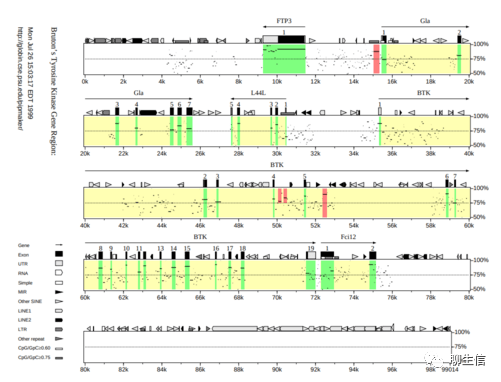
GraphAlignmentViewer
https://github.com/Illumina/GraphAlignmentViewer
Tags: Repeats

PipMaker
http://pipmaker.bx.psu.edu/pipmaker/
Tags: Comparative, Repeat, Annotation
Note: See also MultiPipMaker http://pipmaker.bx.psu.edu/pipmaker/mpm-example/index.html
IGGE
https://github.com/immersivegraphgenomeexplorer/IGGE
Publication: https://www.researchgate.net/profile/Aviv-Elor/publication/354582953_The_Immersive_Graph_Genome_Explorer_Navigating_Genomics_in_Immersive_Virtual_Reality/links/6140c890578238365b0afa3a/The-Immersive-Graph-Genome-Explorer-Navigating-Genomics-in-Immersive-Virtual-Reality.pdf
Tags: Graph, Exotic
Note: Not open source

oxford-plots
https://github.com/jherrero/oxford-plots
Language: R, Perl
Tags: Dotplot
Note: Has nice description of a synteny pipeline here https://github.com/jherrero/oxford-plots/blob/master/examples/pig_X_Y.txt
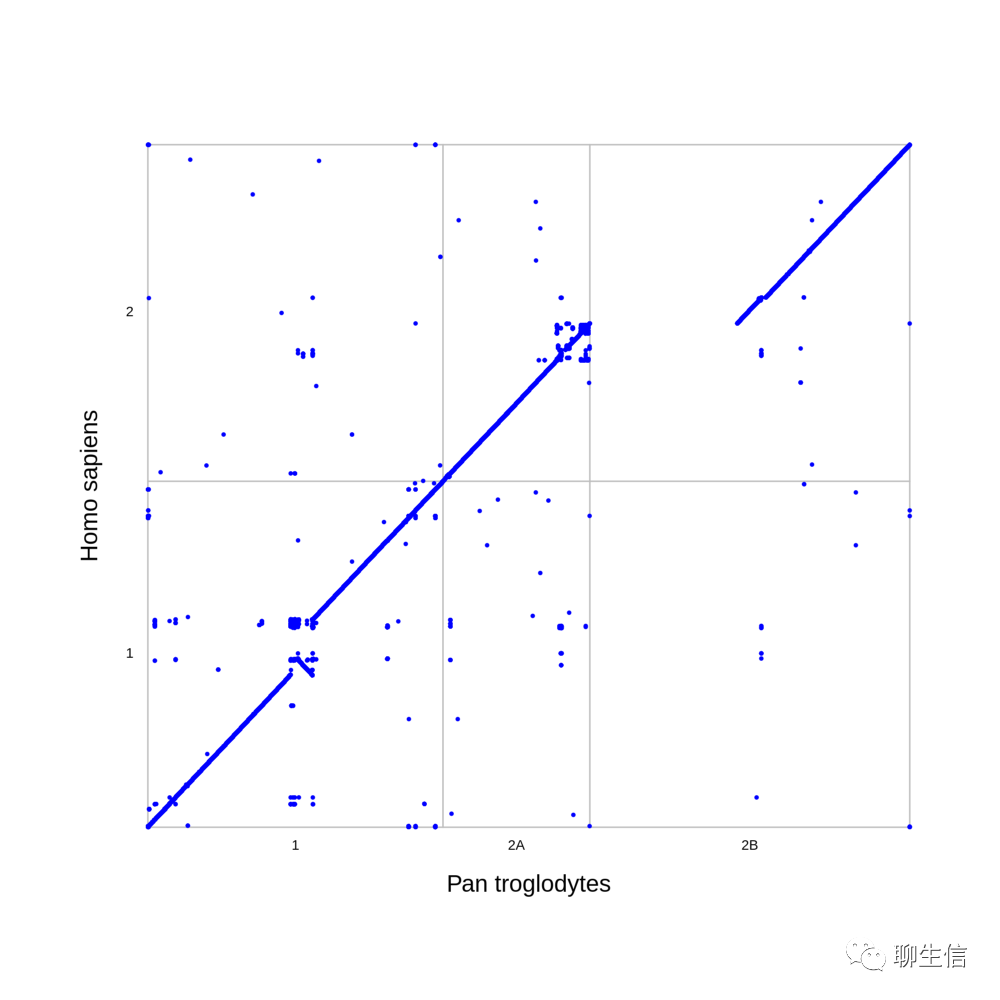
MIRA
http://mira-assembler.sourceforge.net/docs/DefinitiveGuideToMIRA.html
Publication: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.23.7465&rep=rep1&type=pdf
Language: C++
Tags: Assembly, Alignment viewer, Historical
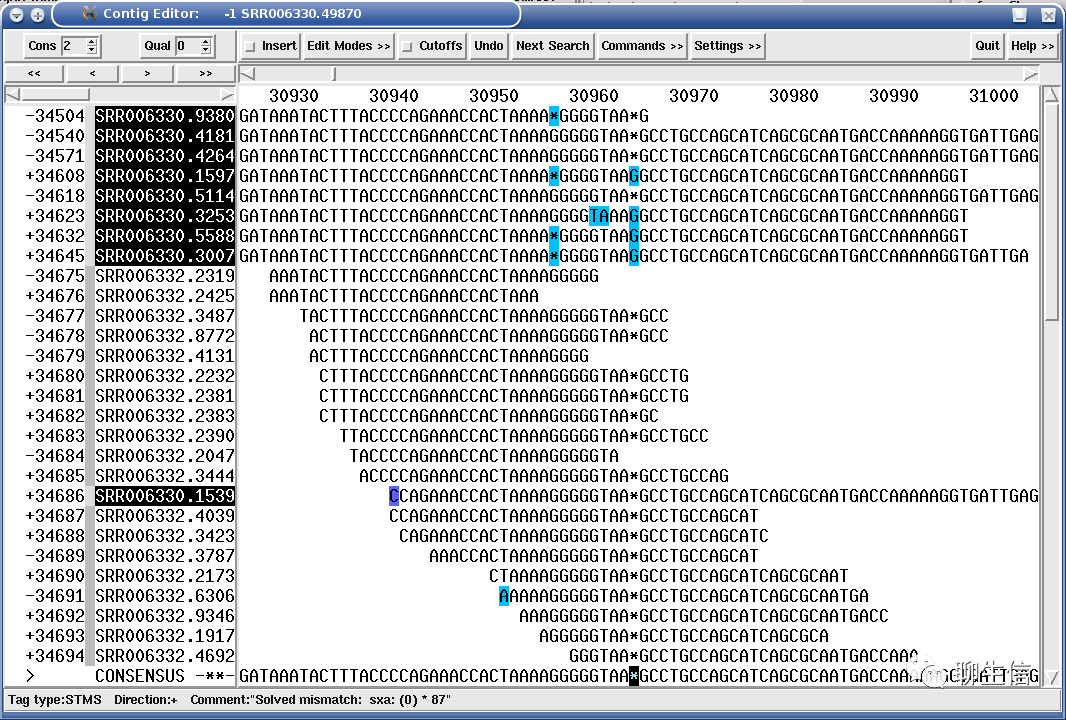
Plotgardener
https://github.com/PhanstielLab/plotgardener/
Publication: https://www.biorxiv.org/content/10.1101/2021.09.08.459338v1
Language: R
Tags: Hi-C, Coverage
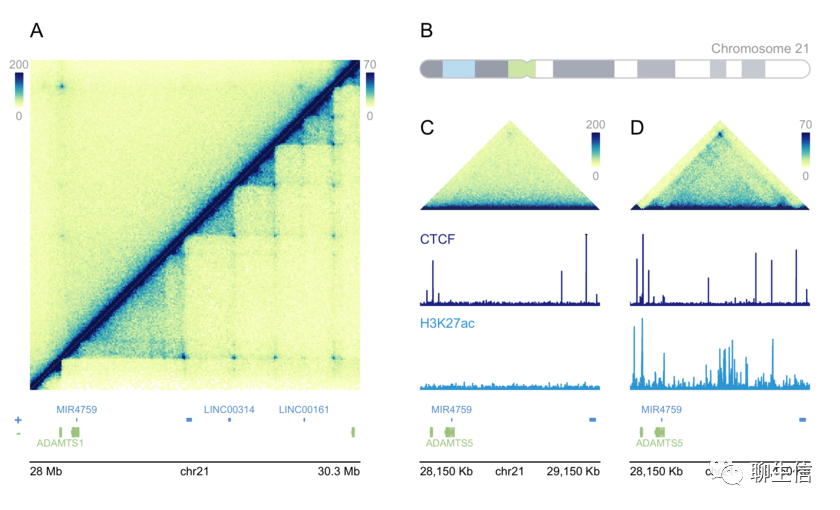
Flash Gviewer
http://gmod.org/wiki/Flashgviewer/
Language: Flash, ActionScript
Tags: Ideogram, Genetic Map, Historical
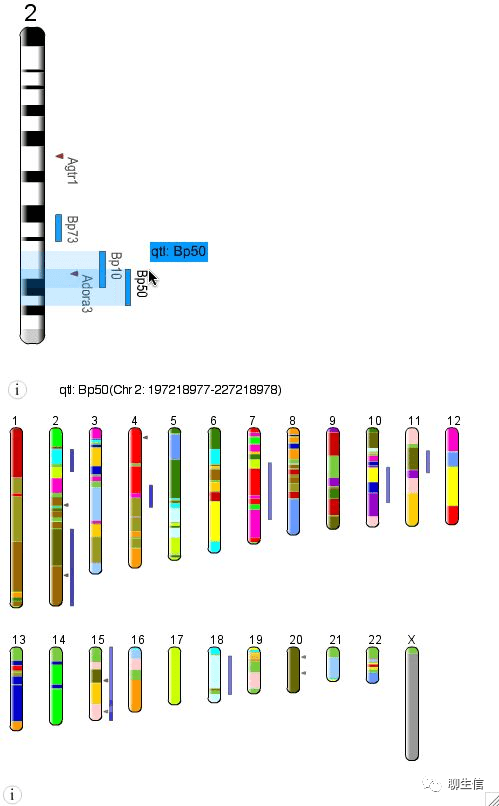
IdeoViz
https://www.bioconductor.org/packages/release/bioc/vignettes/IdeoViz/inst/doc/Vignette.pdf
Language: R
Tags: Ideogram, Quantitative, Splicing
genomegraphs
http://bioconductor.org/packages/2.5/bioc/vignettes/GenomeGraphs/inst/doc/GenomeGraphs.pdf
Language: R
Tags: Ideogram, Quantitative, Splicing
splicejam
https://github.com/jmw86069/splicejam
Language: R
Tags: Splicing, Sashimi
pretzel
https://github.com/plantinformatics/pretzel
Language: JS, D3
Tags: Comparative, Genetic map
wiggleplotr
http://bioconductor.org/packages/devel/bioc/vignettes/wiggleplotr/inst/doc/wiggleplotr.html
Language: R
Tags: Coverage
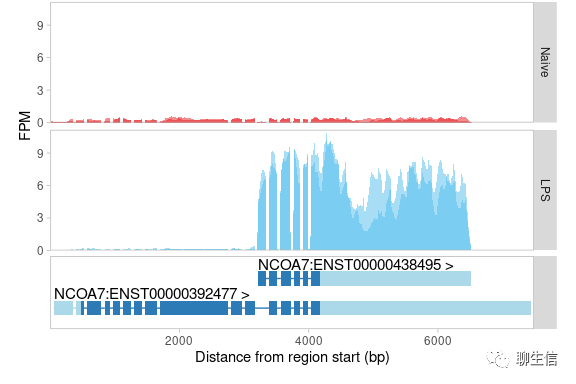
SVPV
https://github.com/VCCRI/SVPV
Publication: https://academic.oup.com/bioinformatics/article/33/13/2032/3056003
Language: Python
Tags: SV, Coverage
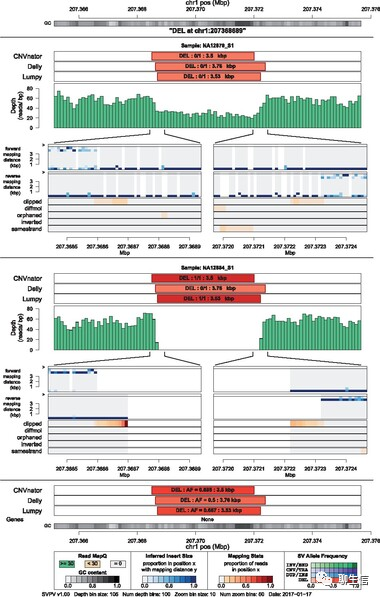
panGraphViewer
https://github.com/TF-Chan-Lab/panGraphViewer
Language: Python
Tags: Graph, Pangenome
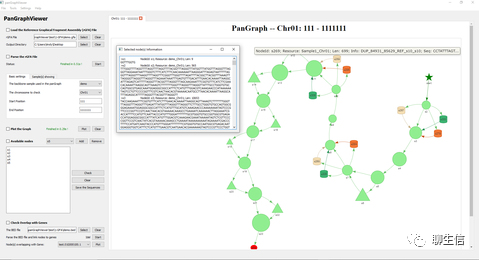
ProViz
http://slim.icr.ac.uk/proviz/index.php
Publication: https://academic.oup.com/nar/article/44/W1/W11/2499309
Language: JS
Tags: MSA, Protein
Note: See also alphafold visualization as tracks http://slim.icr.ac.uk/projects/alphafold?page=alphafold_proviz_homepage
MAVIS
http://mavis.bcgsc.ca/
Language: Python
Tags: Gene fusion, SV
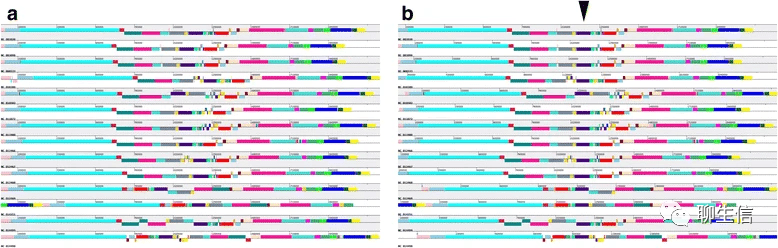
PGAP-X
https://pgapx.zhaopage.com/
Publication: https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-017-4337-7
Language: C++
Tags: Pangenome
Platform: Desktop
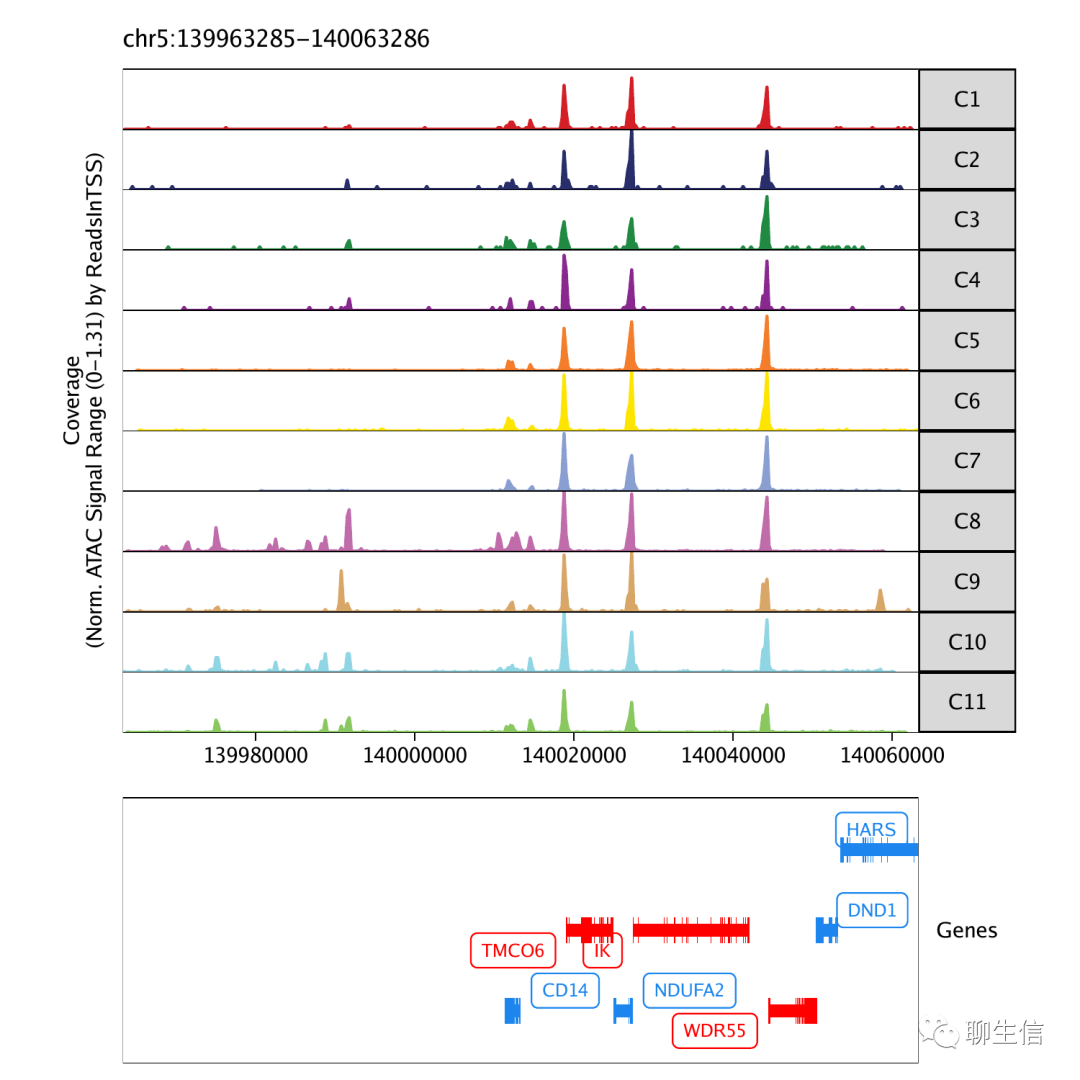
ArchR
https://www.archrproject.com/articles/Articles/tutorial.html
Language: R
Tags: Single cell
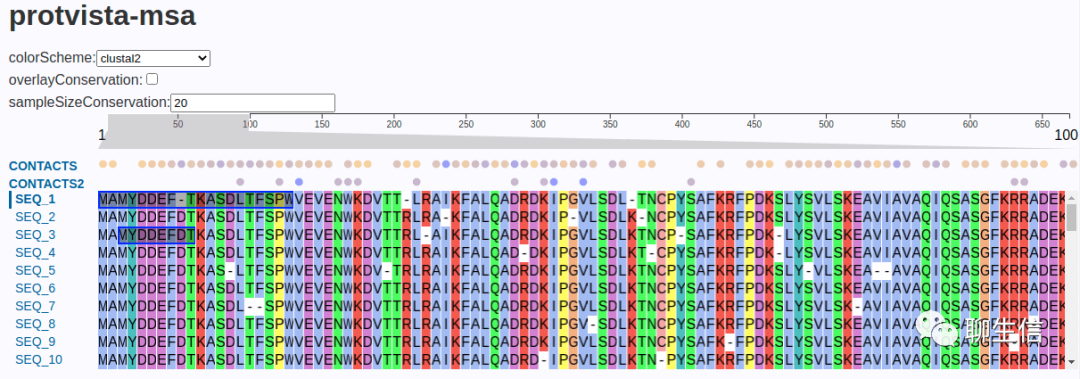
nightingale/protvista
https://ebi-webcomponents.github.io/nightingale/#/msa
Tags: Protein, MSA
Platform: Web
vega
http://vega.archive.ensembl.org/Homo_sapiens/Location/Chromosome?r=6-QBL
Tags: Assembly, Historical, Genetic map
Note: Mentioned in https://www.biorxiv.org/content/10.1101/2021.07.02.450883v1.full.pdf
Platform: Web
Aquaria
http://aquaria.ws/Q9HD67/5i0i/A
Tags: Protein, MSA
Platform: Web
alen
https://github.com/jakobnissen/alen
Language: Rust
Tags: MSA, Text based
NeoLoopFinder
https://github.com/XiaoTaoWang/NeoLoopFinder
Language: Python
Tags: SV, Hi-C
FusionInspector
https://github.com/FusionInspector/FusionInspector/wiki
Language: Perl, JS
Tags: Gene fusion
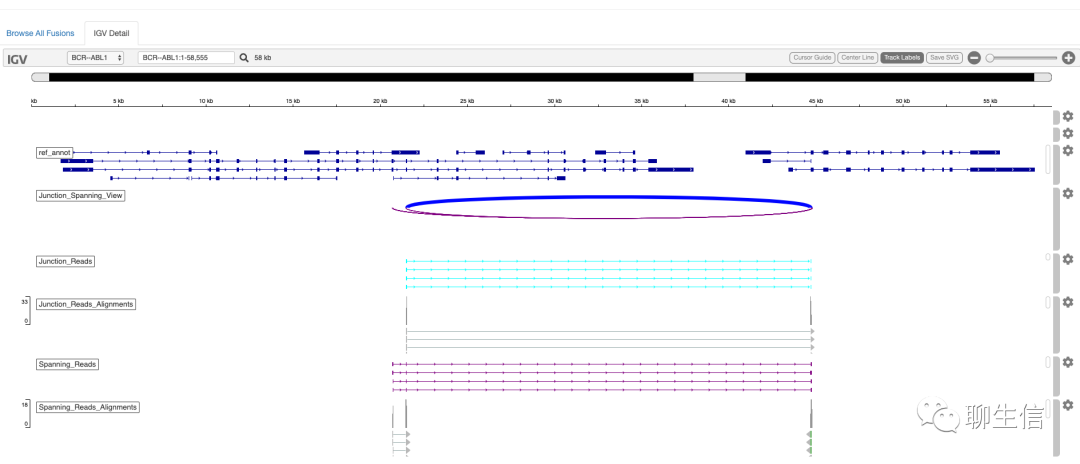
Note: Uses igv.js
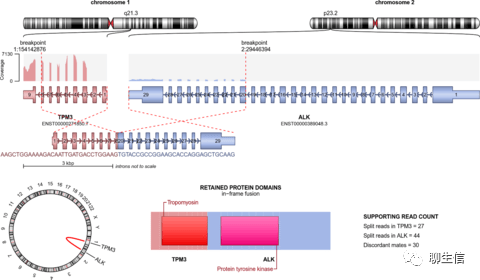
arriba
https://github.com/suhrig/arriba
Publication: https://genome.cshlp.org/content/31/3/448
Language: C++, R
Tags: Gene fusion
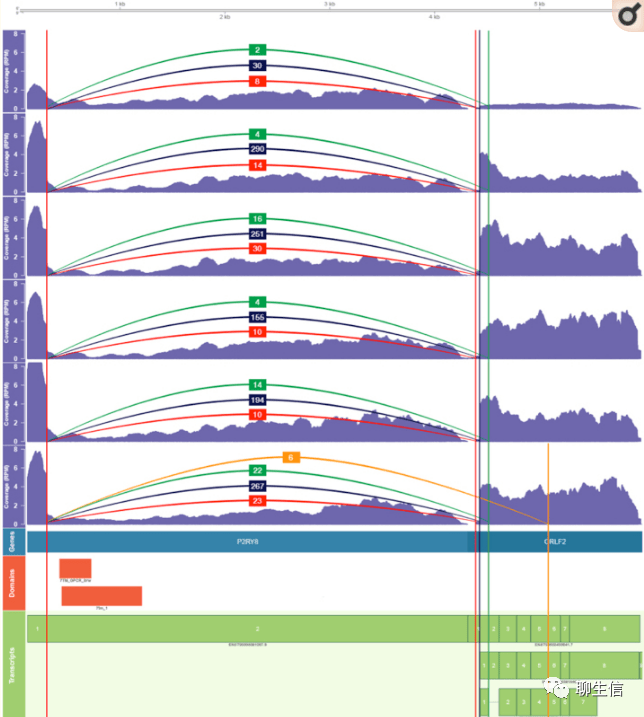
clinker (gene fusion)
https://github.com/Oshlack/Clinker/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6065480/
Language: R
Tags: Gene fusion
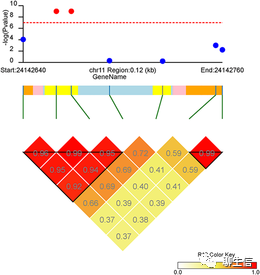
LDBlockShow
https://github.com/BGI-shenzhen/LDBlockShow
Publication: https://academic.oup.com/bib/advance-article-abstract/doi/10.1093/bib/bbaa227/5939575?redirectedFrom=fulltext
Language: C++, Perl
Tags: GWAS, Population
RectChr
https://github.com/BGI-shenzhen/RectChr
Tags: Comparative, Ideogram, Static
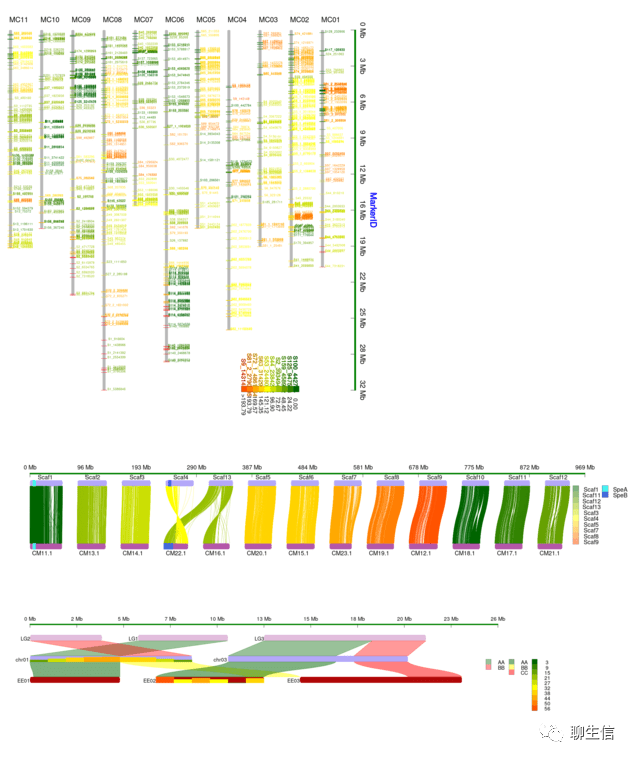
casper
https://github.com/akdess/CaSpER
Publication: https://www.nature.com/articles/s41467-019-13779-x
Language: R
Tags: Single cell, CNV
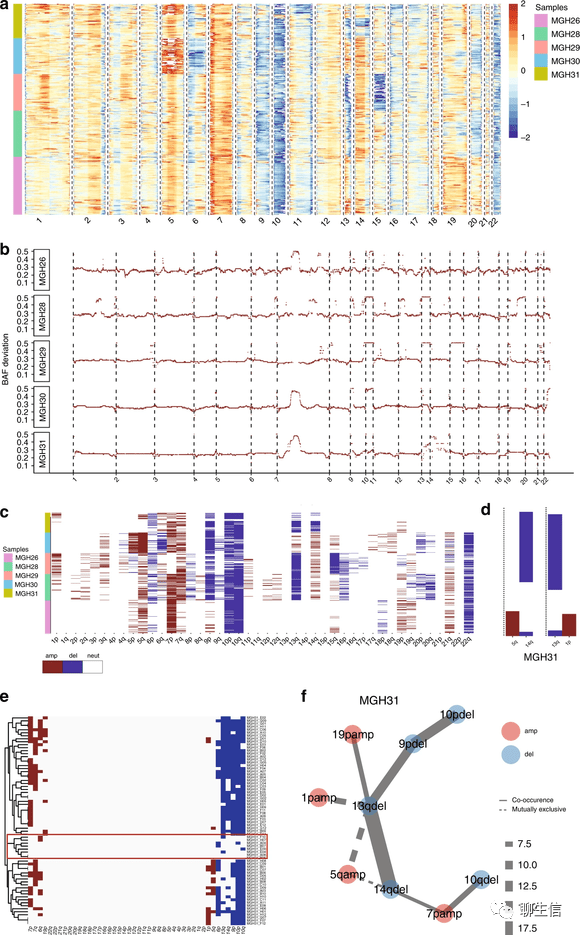
scCNV_heatmap
https://github.com/StefanKurtenbach/scCNV_heatmap
Language: Python
Tags: Single cell, CNV
copykat
https://github.com/navinlabcode/copykat/blob/master/vignettes/copycat-vignettes.pdf
Language: R
Tags: Single cell, CNV
alignfigR
https://cran.r-project.org/web/packages/alignfigR/vignettes/my-vignette.html
Language: R
Tags: MSA
copynumber
https://www.bioconductor.org/packages/release/bioc/vignettes/copynumber/inst/doc/copynumber.pdf
Language: R
Tags: CNV
SWAV
http://swav.popgenetics.net/
Publication: https://www.nature.com/articles/s41598-019-57038-x
Tags: Population, Deadlink
methylartist
https://github.com/adamewing/methylartist
Language: Python
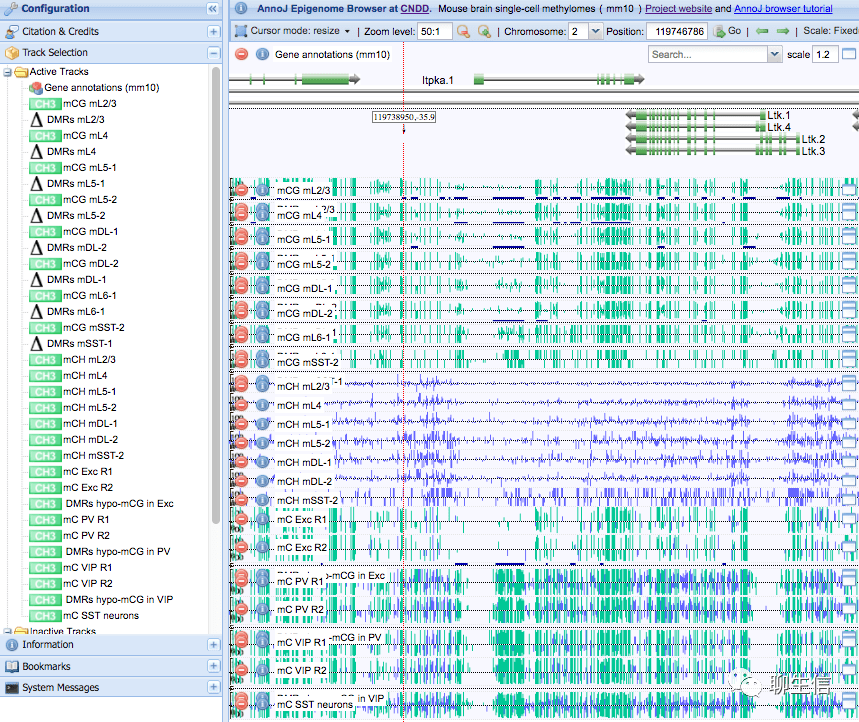
AnnoJ
https://brainome.ucsd.edu/howto_annoj.html
Language: JS
Tags: Methylation
Note: See list of instances of the browser here https://ecker.salk.edu/genome-browser/
Github: https://github.com/mukamel-lab/annoj_cndd
Platform: Web
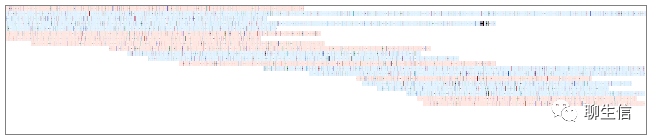
udon
https://github.com/ocxtal/udon
Language: Rust
Tags: Alignment viewer
Note: Uses an advanced data structure for pileup, so visually a basic example but likely just a small demo
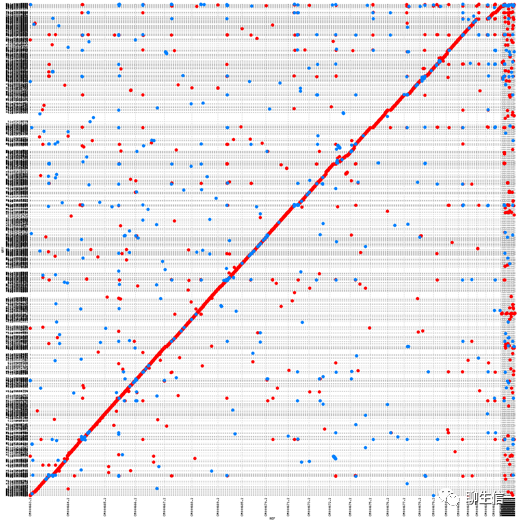
MashMap
https://github.com/marbl/MashMap/blob/master/scripts/generateDotPlot
Tags: Dotplot
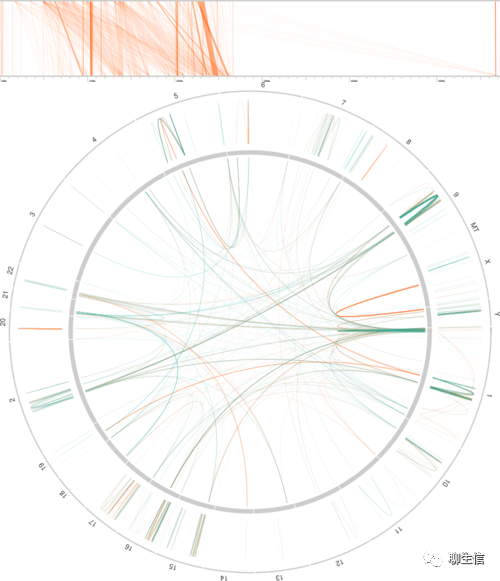
asgart
https://github.com/delehef/asgart
Publication: https://academic.oup.com/bioinformatics/article/34/16/2708/4948616
Language: Rust
Tags: SV, Ideogram, Circular, Linear
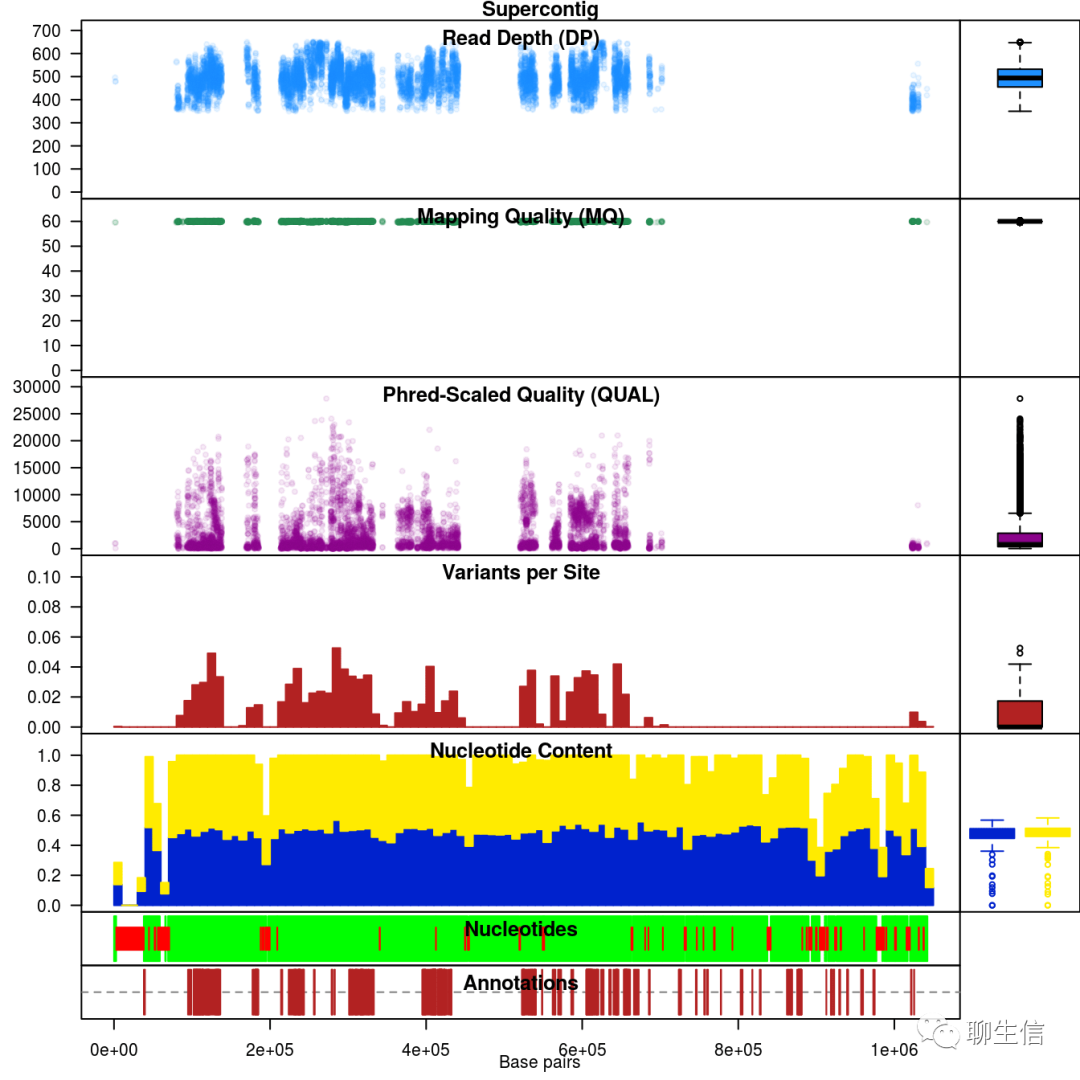
vcfR
https://knausb.github.io/vcfR_documentation/
Language: R
Tags: QC, Population, Coverage
Note: image shows chromoqc output
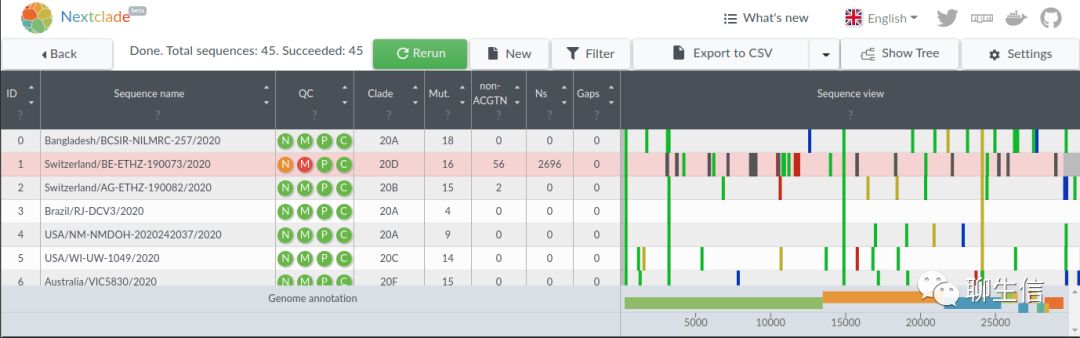
Nextclade
https://clades.nextstrain.org/
Language: JS, React, D3
Tags: Phylogenetics
Note: Related usage on nextstrain app also
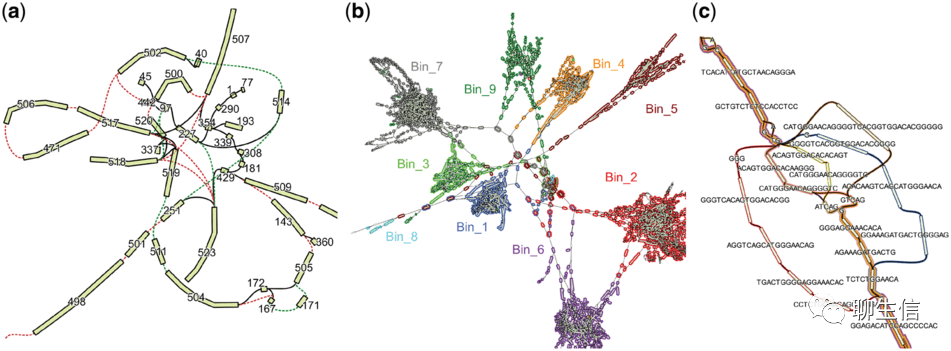
GfaViz
Publication: https://academic.oup.com/bioinformatics/article/35/16/2853/5267826
Language: C++
Tags: Graph
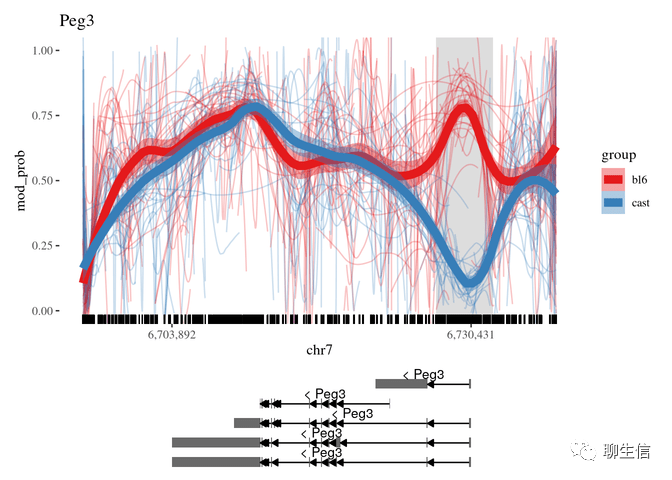
NanoMethViz
http://www.bioconductor.org/packages/release/bioc/html/NanoMethViz.html
Publication: https://www.biorxiv.org/content/10.1101/2021.01.18.426757v3
Language: R
Tags: Methylation
genevisR
https://bioconductor.org/packages/release/bioc/vignettes/GenVisR/inst/doc/Intro.html
Language: R
Tags: CNV
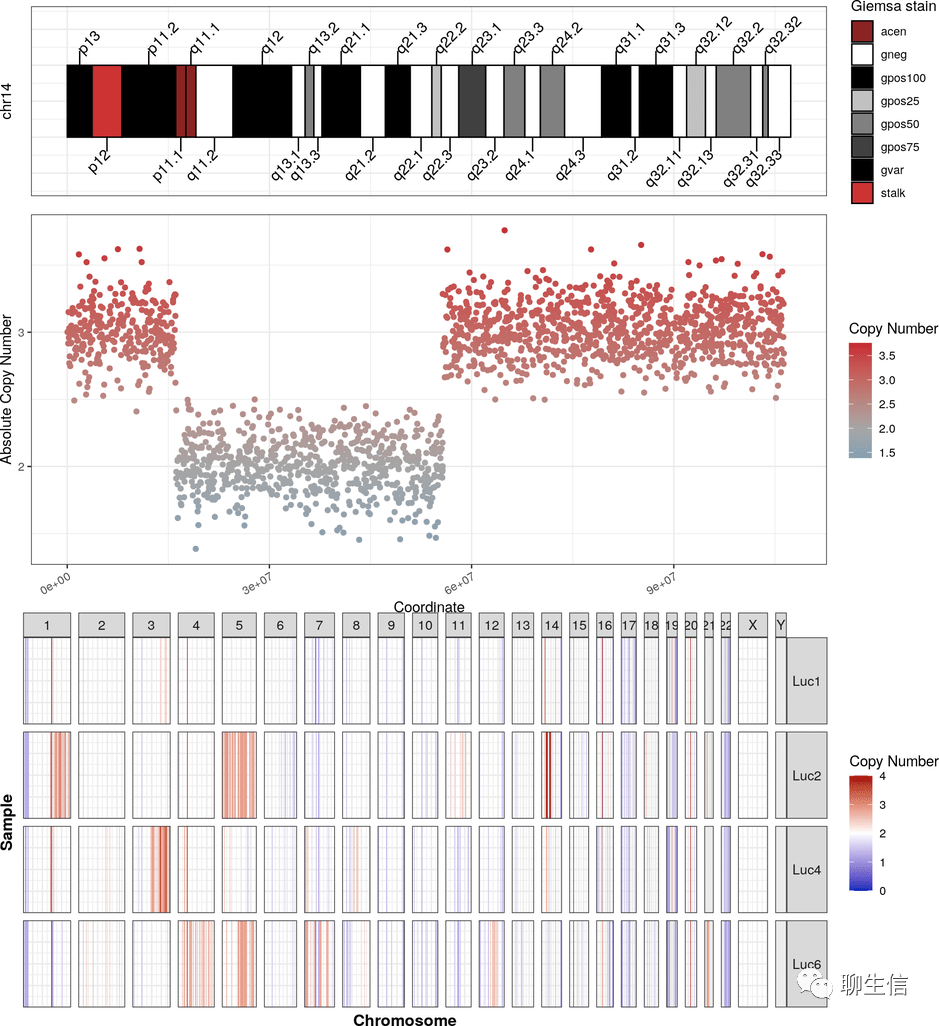
ALVIS (chimeric alignment viewer)
https://github.com/SR-Martin/alvis
Publication: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-021-04056-0
Tags: QC, Assembly
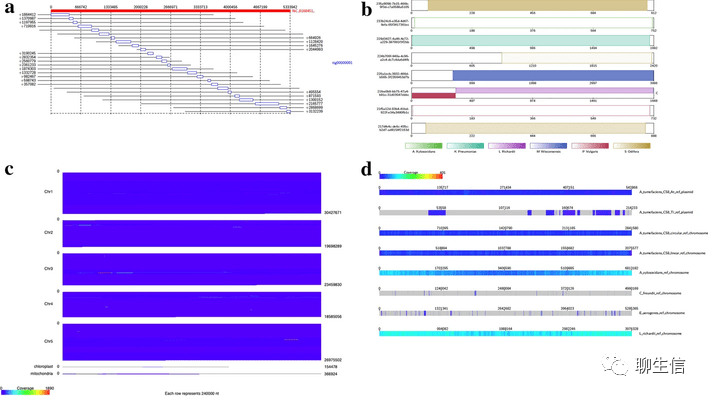
ALVIS (MSA viewer)
https://www.ebi.ac.uk/research/goldman/software/alvis
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4856975/
Tags: MSA
Note: Introduces sequence bundles concept, also see web app version https://www.ebi.ac.uk/goldman-srv/sequencebundles/
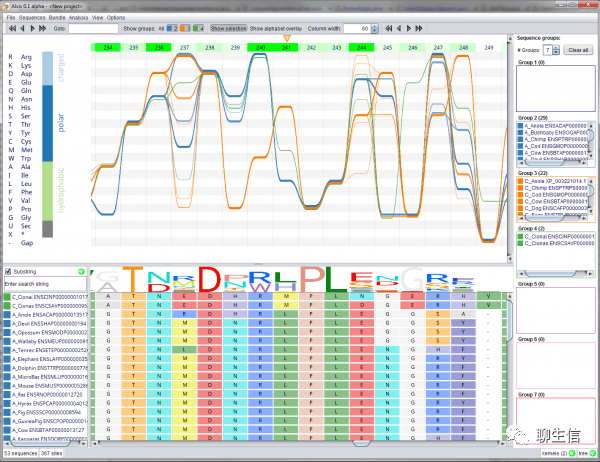
mocha
https://github.com/freeseek/mocha#plot-results
Language: C, WDL, R
Tags: CNV
Note: Has a full analysis pipeline associated with end result visualizations
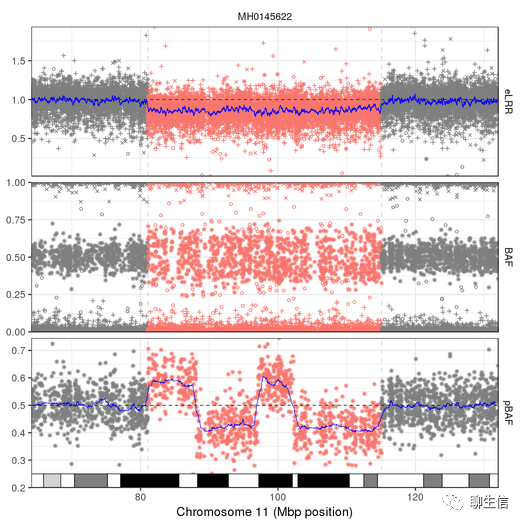
bctools cnv
https://samtools.github.io/bcftools/howtos/cnv-calling.html
Tags: CNV
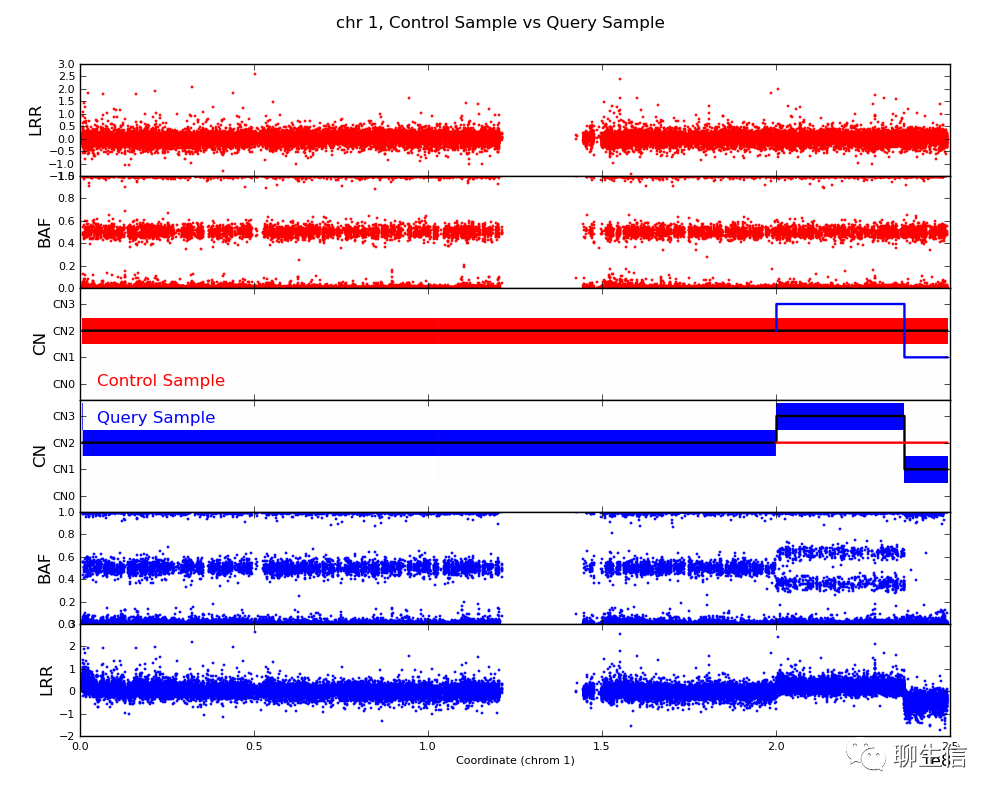
bcftools roh
https://samtools.github.io/bcftools/howtos/roh-calling.html
Language: Python
Tags: Static
Note: Plots runs of homozygosity, has interactive plot-roh.py helper too
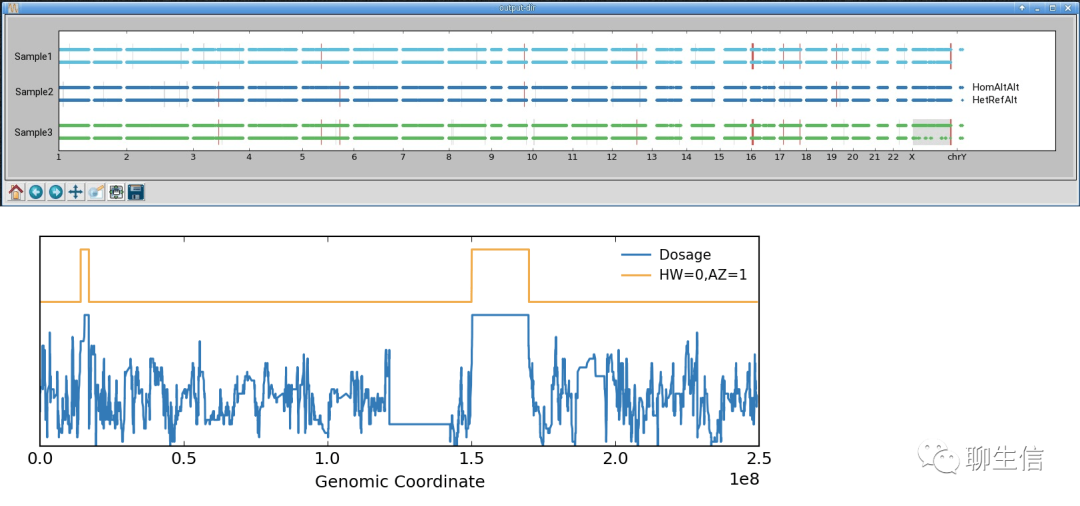
Internet Contig Explorer
https://www.bcgsc.ca/resources/software/ice
Publication: https://genome.cshlp.org/content/13/6a/1244.long
Tags: Historical
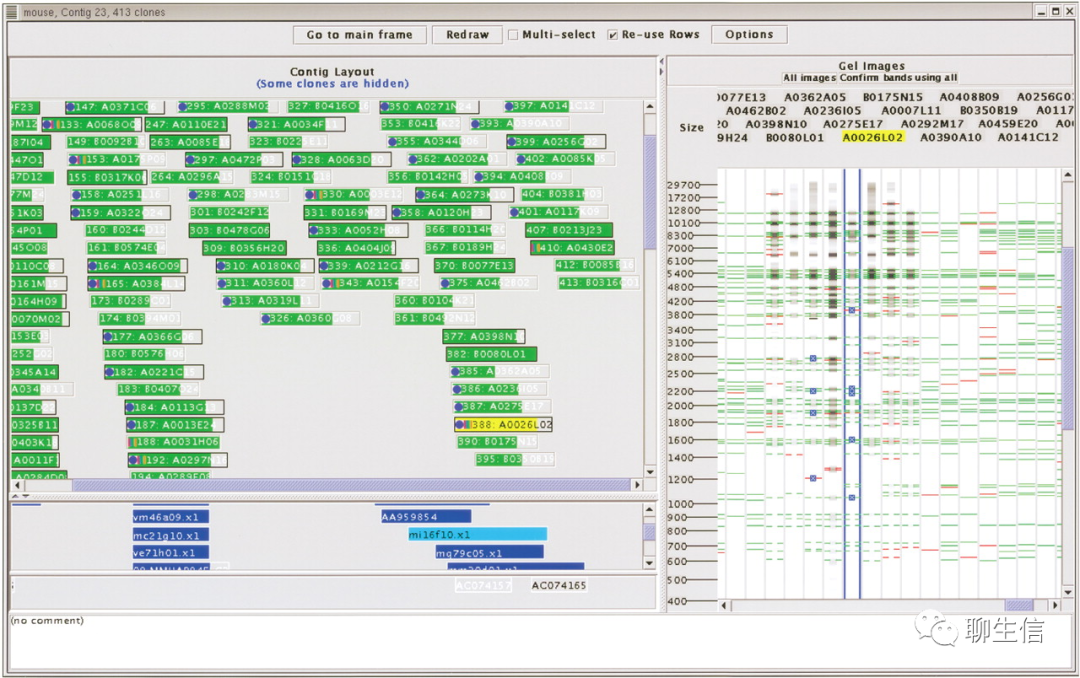
spinteny
https://github.com/skinner/spinteny
Tags: Exotic, Comparative

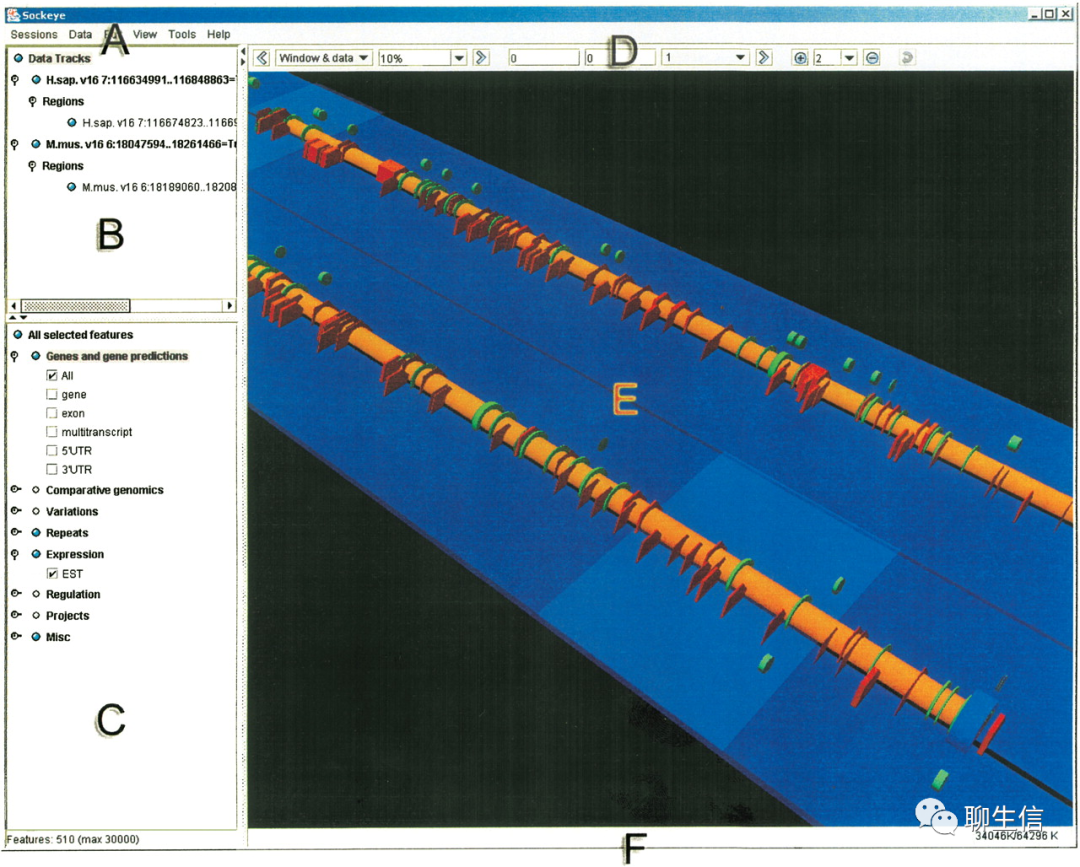
sockeye
https://www.bcgsc.ca/resources/software/sockeye
Publication: https://genome.cshlp.org/content/14/5/956.full
Tags: Exotic, Comparative
SMRT View
http://files.pacb.com/software/smrtanalysis/2.3.0/doc/smrtview/help/Webhelp/App_View_Epipro.htm
Tags: SV
Github: https://github.com/PacificBiosciences/DevNet/wiki/SMRT-View
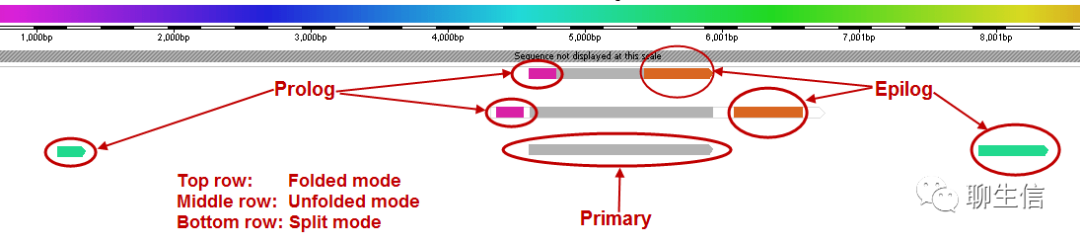
PURPLE
https://github.com/hartwigmedical/hmftools/blob/master/purity-ploidy-estimator/README.md
Tags: CNV, Cancer
Note: intermutation-distance rainfall plots/katagesis plots are not technically in genomic coordinates, but do indicate genomic cluster of variatnts
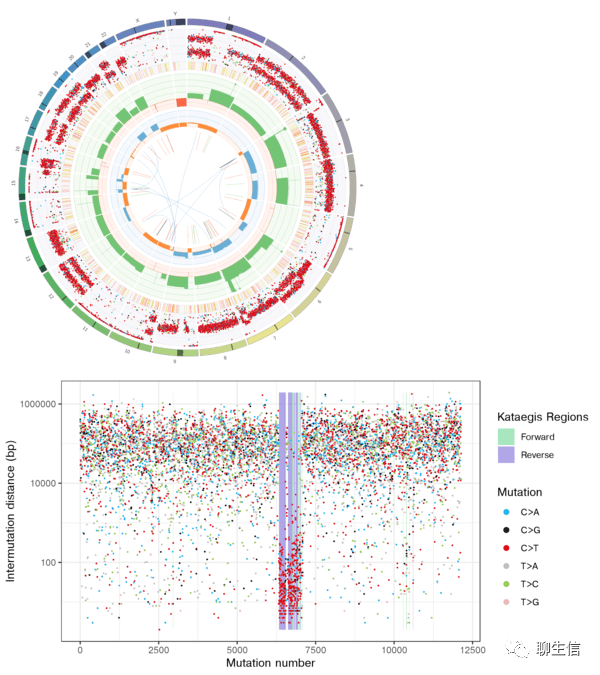
LINX
https://github.com/hartwigmedical/hmftools/blob/master/sv-linx/README_VIS.md
Tags: Graph, SV, Breakends, Circular, Linear, Gene fusion, Cancer
Note: part of the PURPLE/GRIDSS/LINX pipeline
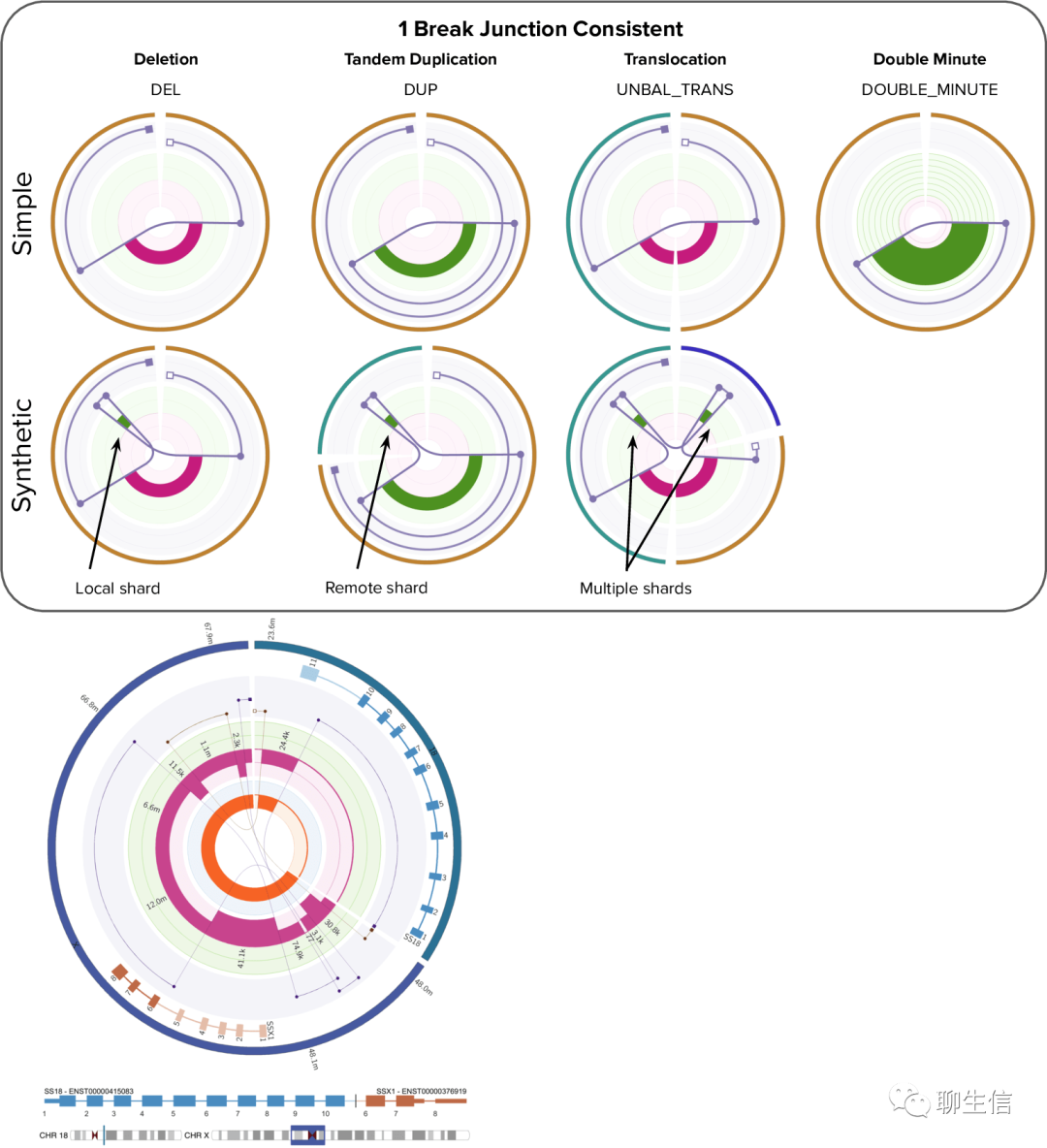
Shasta
https://chanzuckerberg.github.io/shasta/ComputationalMethods.html#ReadGraph
Language: C++
Tags: Graph
Note: Uses graphviz
Platform: Web, Localhost, CLI
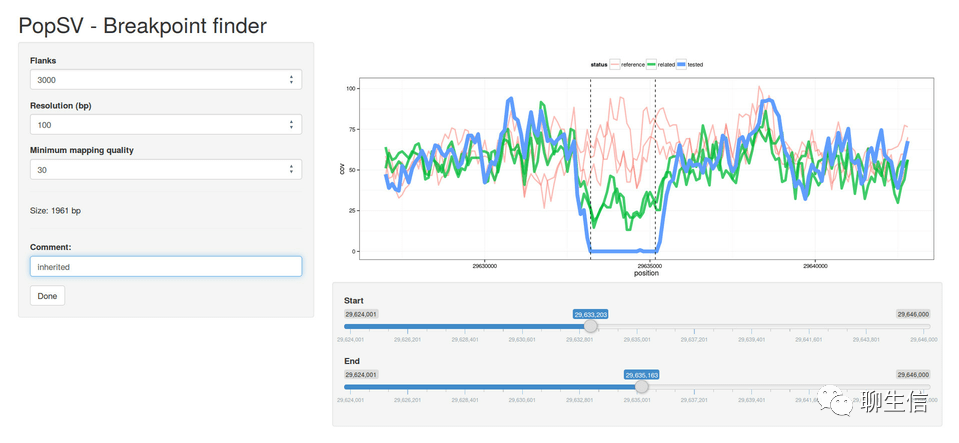
PopSV
https://github.com/jmonlong/PopSV/blob/master/3-Visualization.md
Language: R
Tags: Population, SV
Platform: Web
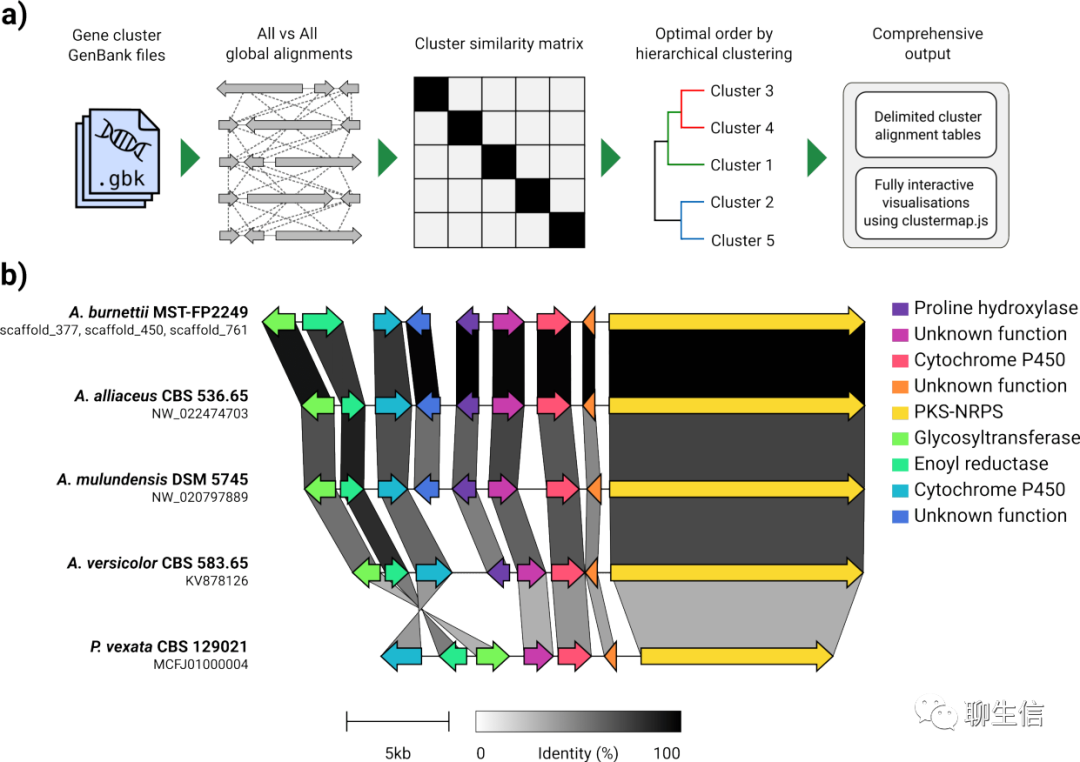
clinker
https://github.com/gamcil/clinker
Publication: https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btab007/6103786
Language: JS
Tags: Comparative, Gene order
Platform: Web
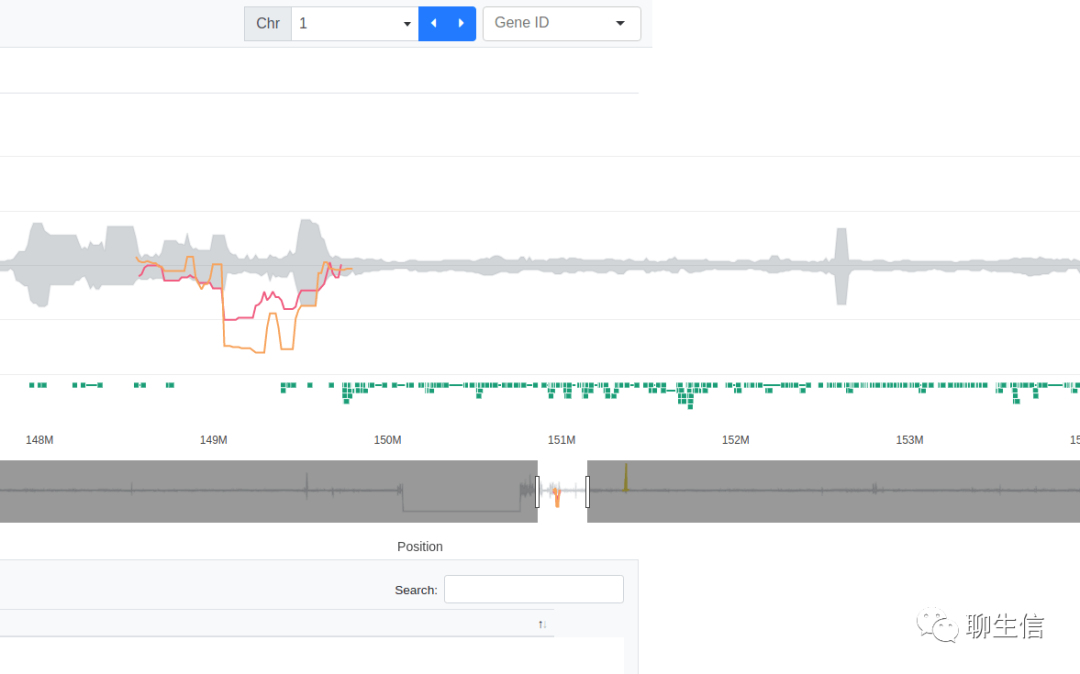
covviz
https://brwnj.github.io/covviz/
Language: JS
Tags: CNV, Coverage, Population
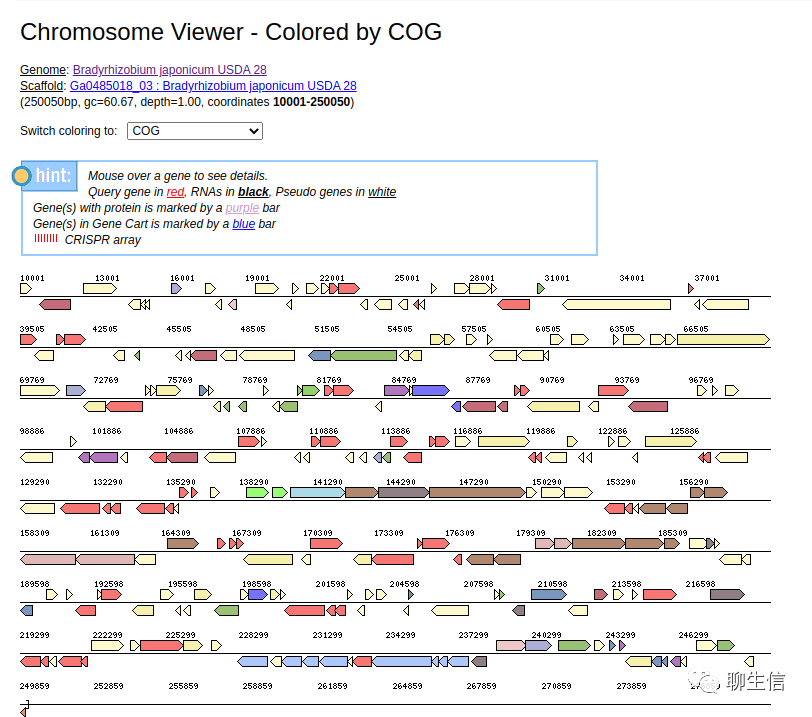
JGI/IMG
https://img.jgi.doe.gov/
Tags: Dotplot, Linear, Microbiology
Note: Dotplot use MUMmer
Platform: Web, Silo
FastANI
https://github.com/ParBLiSS/FastANI
Language: C++
Tags: Comparative, Microbiology
Gview
https://server.gview.ca/examples
Tags: Circular, Linear, Pangenome, Microbiology
Platform: Web
LinearDisplay.pl
https://github.com/JCVenterInstitute/LinearDisplay
Language: Perl
Tags: Comparative
Platform: CLI

REPAVER
https://gitlab.com/gringer/bioinfscripts/
Publication: https://www.researchgate.net/publication/346507956_REPAVER_Paving_the_Way_for_Repeat_Discovery_in_Complex_Genomes?channel=doi&linkId=5fc58ba4a6fdcce95268fe60&showFulltext=true
Language: R, C++
Tags: Repeats, Dotplot, Static, Linear
Platform: CLI
Dotter
https://sonnhammer.sbc.su.se/Dotter.html
Publication: https://pubmed.ncbi.nlm.nih.gov/8566757/
Tags: Dotplot
Platform: CLI
VIVA
https://www.biorxiv.org/content/10.1101/589879v1.full.pdf
Tags: Heatmap, Population, Variants
Platform: Web
UCSC Xena
https://xena.ucsc.edu/
Tags: Heatmap, Population
Platform: Web
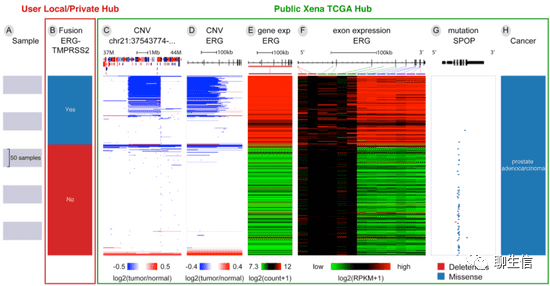
pViz
https://github.com/Genentech/pviz
Tags: Protein
Platform: Web
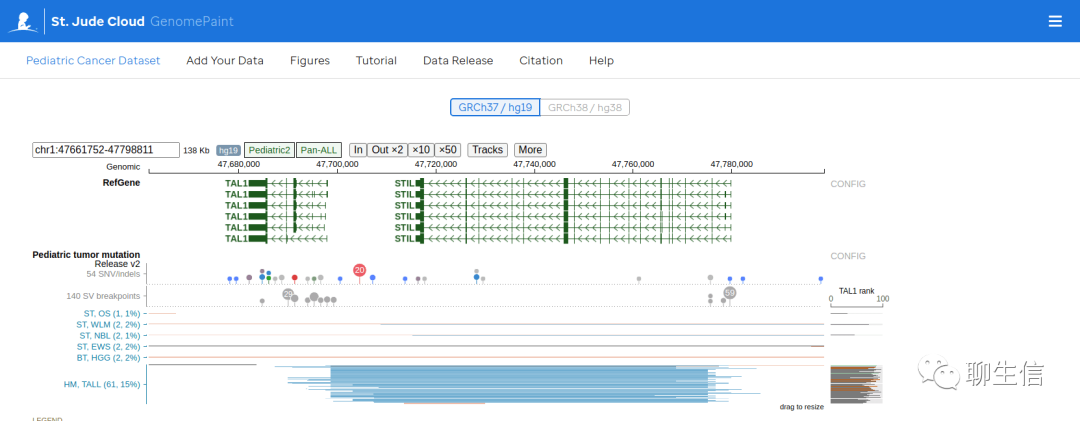
GenomePaint
https://genomepaint.stjude.cloud/
Publication: https://www.cell.com/cancer-cell/fulltext/S1535-6108(20)30659-0
Tags: CNV, Linear, Cancer, Lollipops
Note: See also gallery here https://genomepaint.stjude.cloud/#figures
Platform: Web
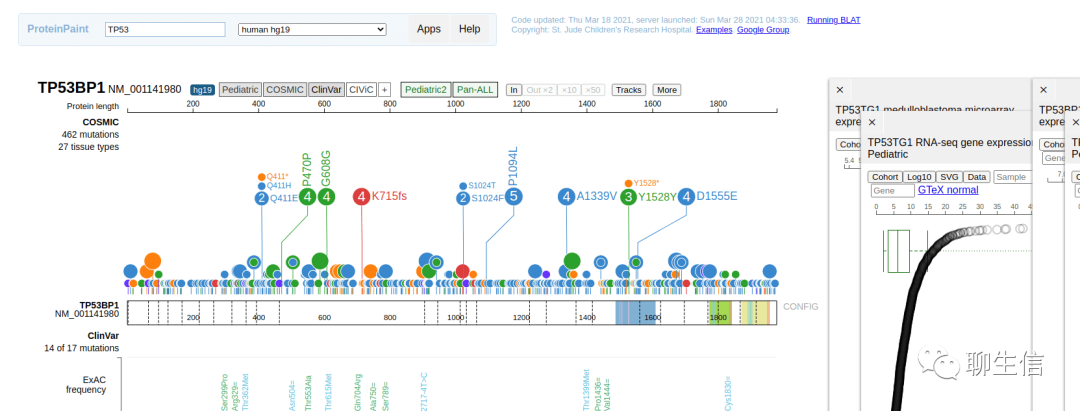
ProteinPaint
https://pecan.stjude.cloud/proteinpaint
Publication: https://www.nature.com/articles/ng.3466
Tags: Protein, Cancer, Lollipops
Platform: Web
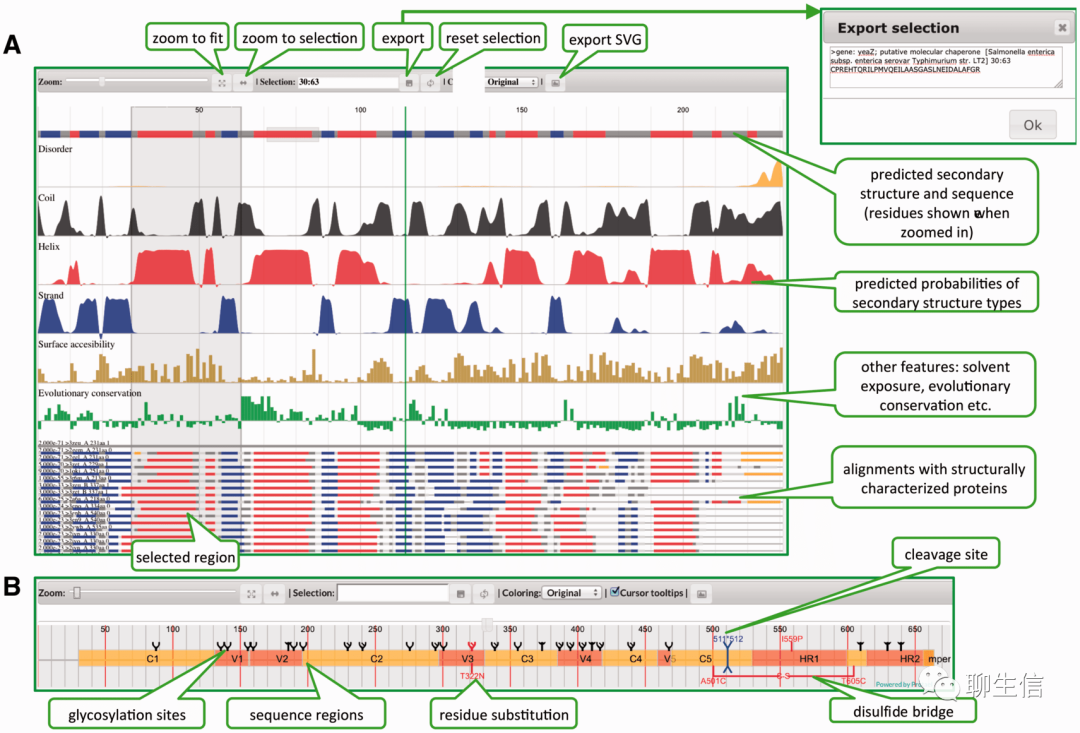
Protael
http://sanshu.github.io/protaelweb/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5963358/
Language: JS
Tags: Protein, MSA
Platform: Web
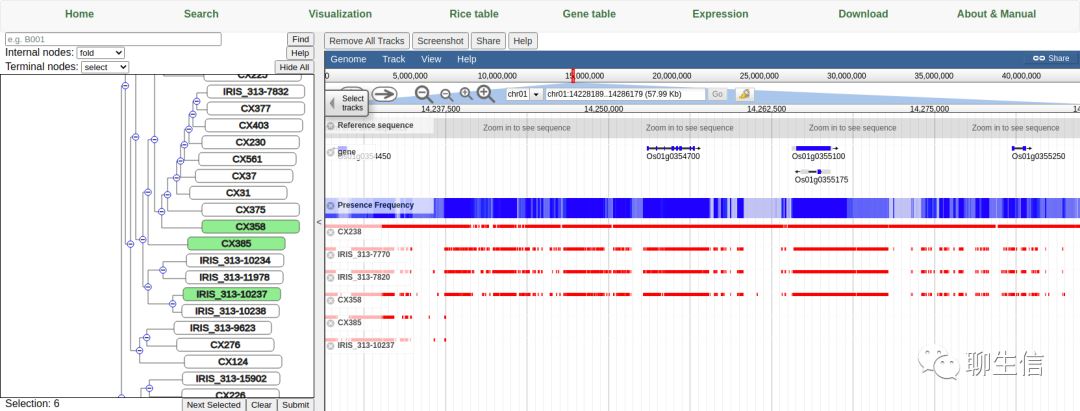
RPAN (3kricedb)
http://cgm.sjtu.edu.cn/3kricedb/visualization/?tracks=DNA%2Cgene%2CPF
Tags: Pangenome, JBrowse integration
Platform: Web
Panache
https://github.com/SouthGreenPlatform/panache
Tags: Pangenome
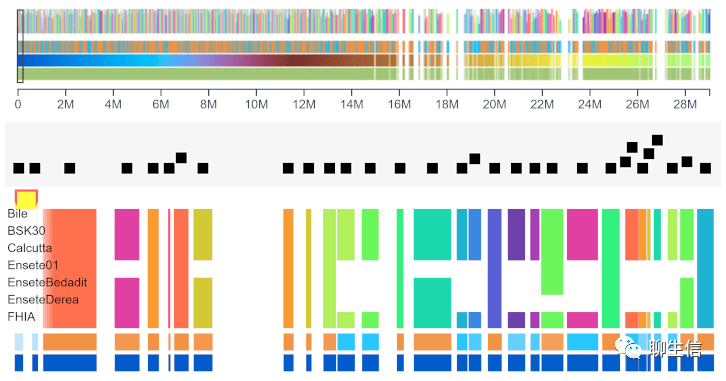
Panacaea
https://github.com/JCVenterInstitute/PanACEA
Tags: Pangenome
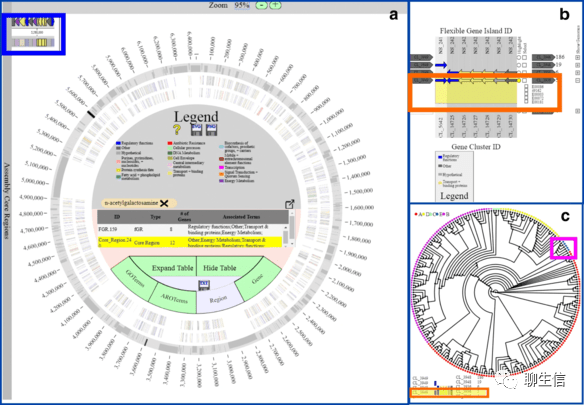
Pan-Tetris
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4547177/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4547177/
Tags: Pangenome
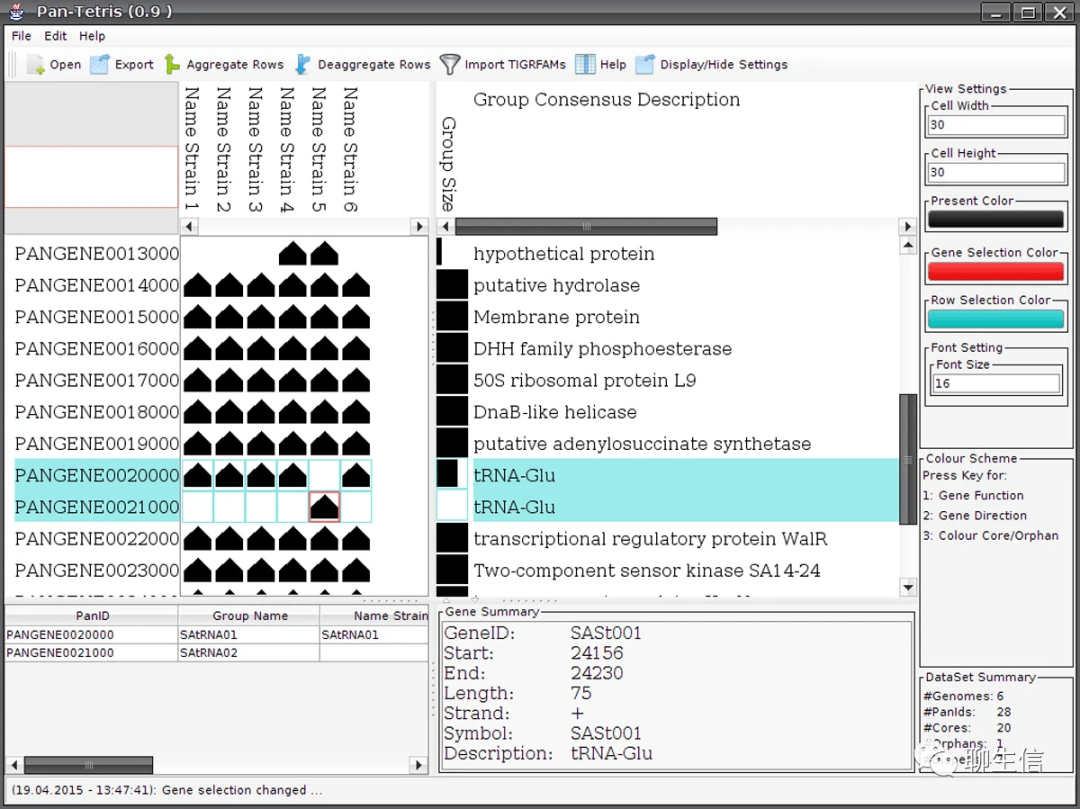
iGenomics
https://www.biorxiv.org/content/10.1101/2020.02.11.944132v1
Tags: Mobile
UCSC Genome Graphs
http://genome.ucsc.edu/cgi-bin/hgGenome
Tags: Ideogram
Platform: Web, Form, Silo

NCBI Genome Decoration
https://www.ncbi.nlm.nih.gov/genome/tools/gdp/
Tags: Ideogram
Platform: Web, Form, Silo
KaryotypeSVG
https://github.com/andreasprlic/karyotypeSVG
Language: JS, SVG
Tags: Ideogram
Platform: Web
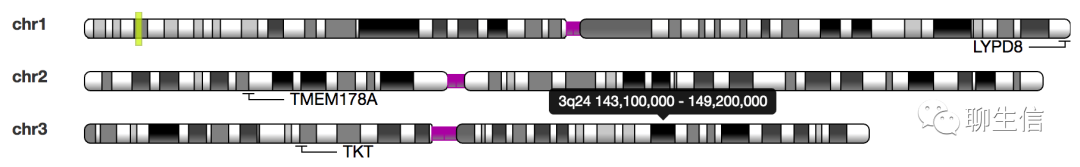
karyoploteR
https://github.com/bernatgel/karyoploteR
Language: R
Tags: Ideogram, Static

Ideogram
https://github.com/RCollins13/HumanIdiogramLibrary
Language: SVG
Tags: Ideogram, Static
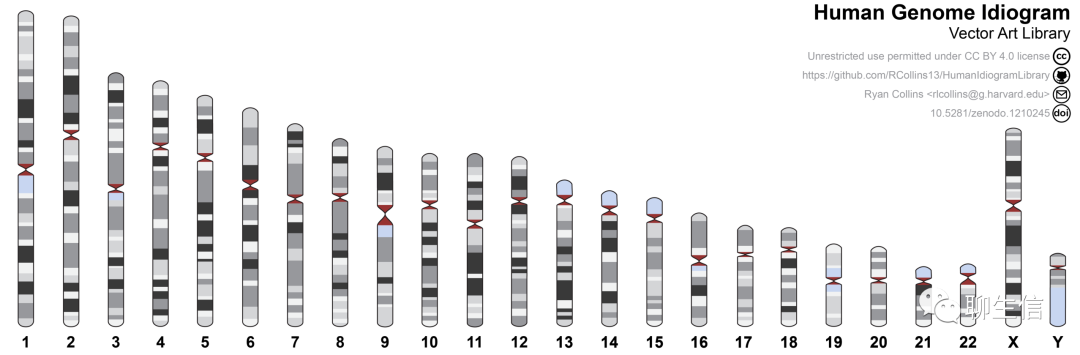
eweitz/Ideogram
https://github.com/eweitz/ideogram
Language: JS
Tags: Ideogram
Platform: Web
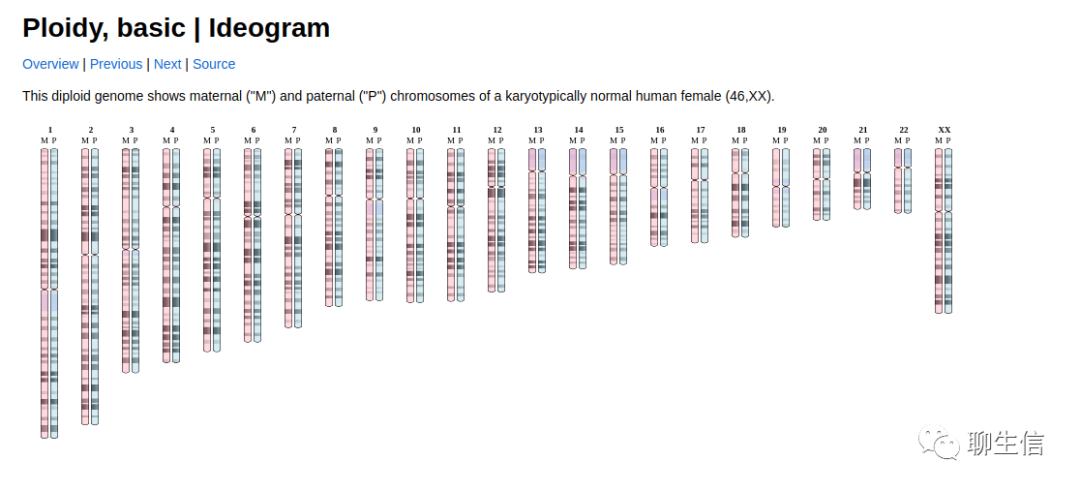
chromoMap
https://www.biorxiv.org/content/10.1101/605600v1.full.pdf
Language: R
Tags: Ideogram, Static
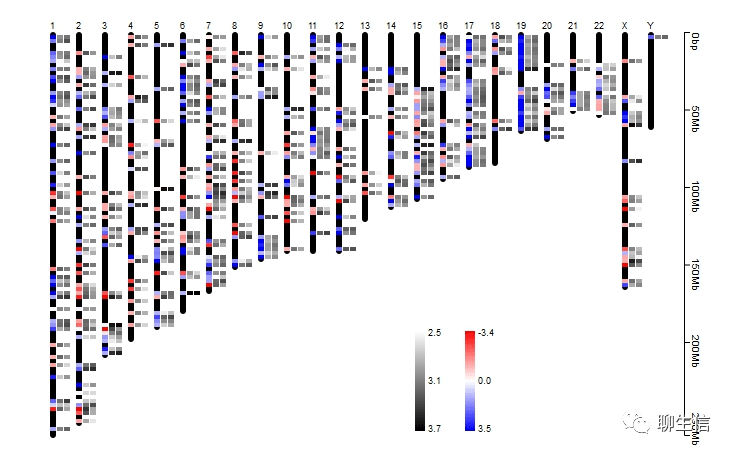
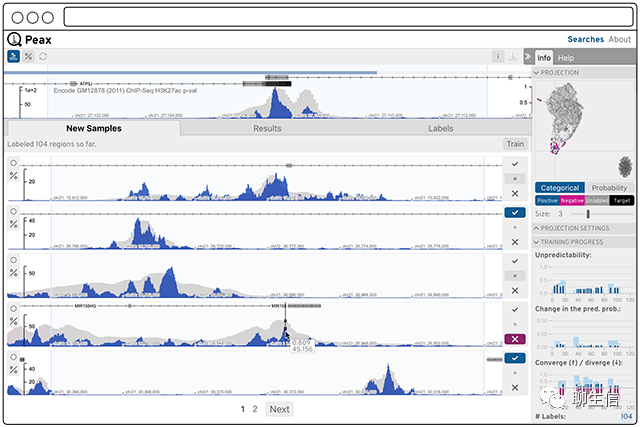
Peax
https://github.com/Novartis/peax
Language: JS
Tags: Epigenomics, Higlass integration
Platform: Web
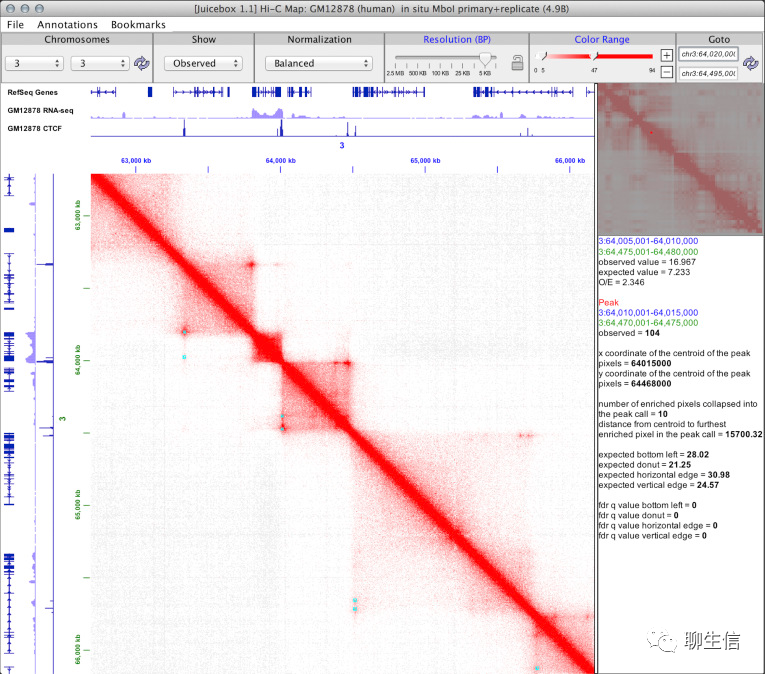
Juicebox.js
http://www.igv.org/doc/juiceboxjs.html
Publication: https://www.cell.com/fulltext/S2405-4712(18)30001-2
Language: JS
Tags: Epigenomics, Hi-C
Platform: Web
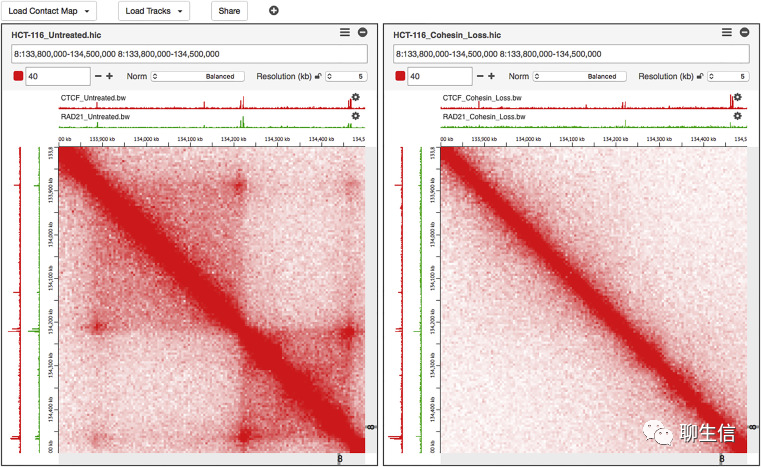
Juicebox
https://github.com/aidenlab/Juicebox
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5596920/
Language: Java
Tags: Epigenomics, Hi-C
Platform: Desktop
HiPiler
http://hipiler.higlass.io
Tags: Epigenomics, Hi-C, Higlass integration
Platform: Web

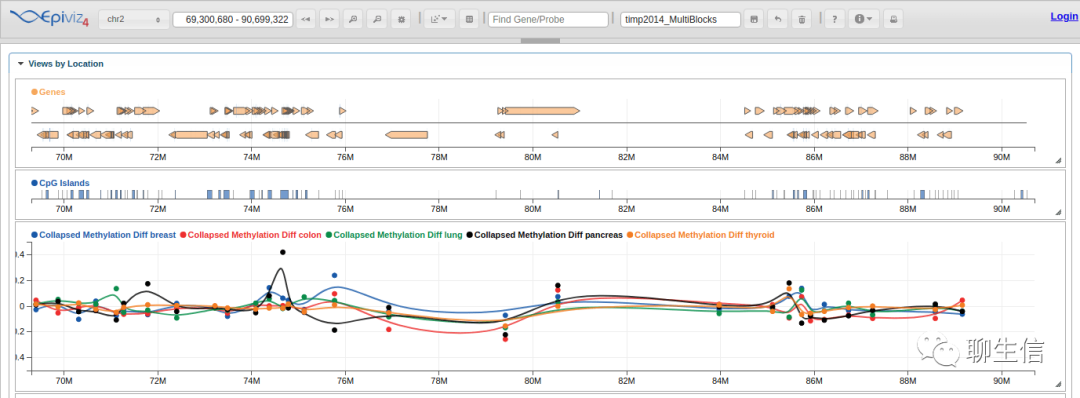
Epiviz
https://epiviz.github.io/
Publication: https://www.biorxiv.org/content/10.1101/865295v1.full.pdf
Language: JS, R, D3
Tags: Epigenomics
Platform: Web
Epilogos
https://epilogos.altius.org/
Tags: Epigenomics, Higlass integration
Platform: Web

cisGenome Browser
https://jhui2014.github.io/browser/screenshots.html
Language: C
Tags: Epigenomics
Platform: Desktop
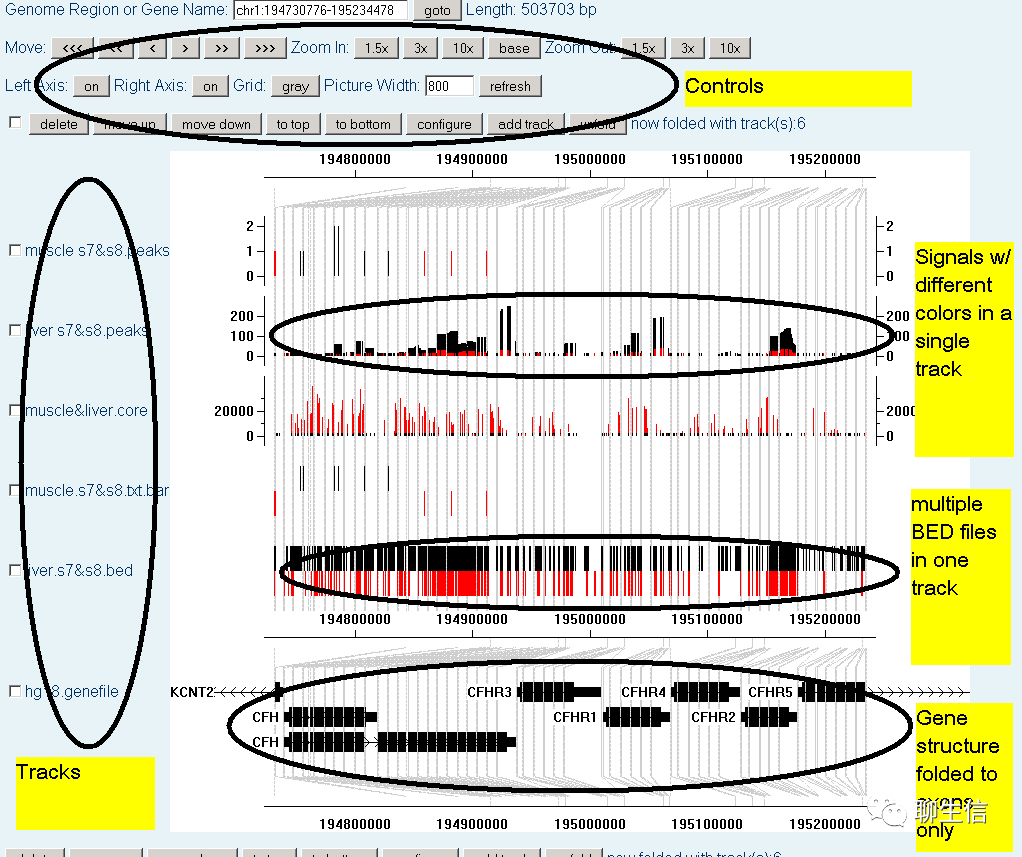
Chipster
https://chipster.csc.fi/
Language: Java
Tags: Epigenomics, CNV
Note: Copy number tutorial https://chipster.csc.fi/manual/cn-tutorial.pdf
Platform: Desktop
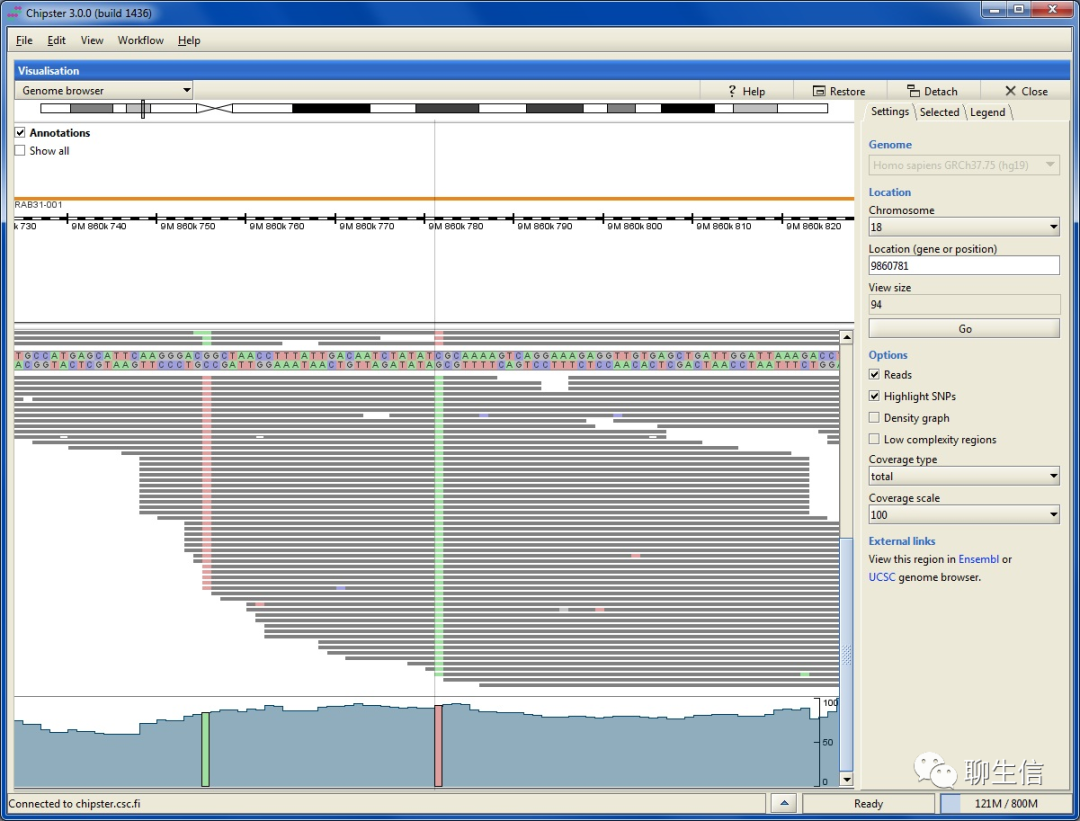
Chip Monk
http://www.bioinformatics.babraham.ac.uk/projects/chipmonk/
Language: Java
Tags: Epigenomics
Note: Also see SeqMonk https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/
Platform: Desktop
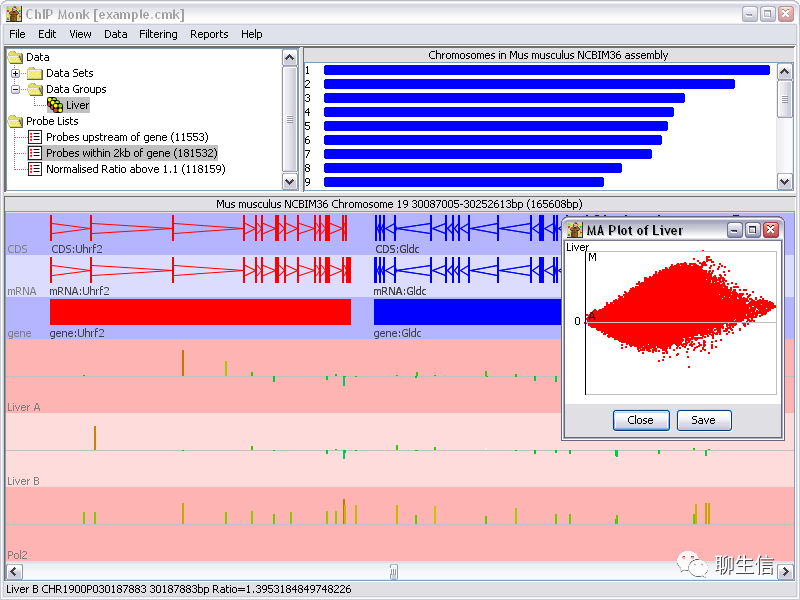
syntenyPlotByR
https://github.com/shingocat/syntenyPlotByR
Language: R
Tags: Dotplot, Static
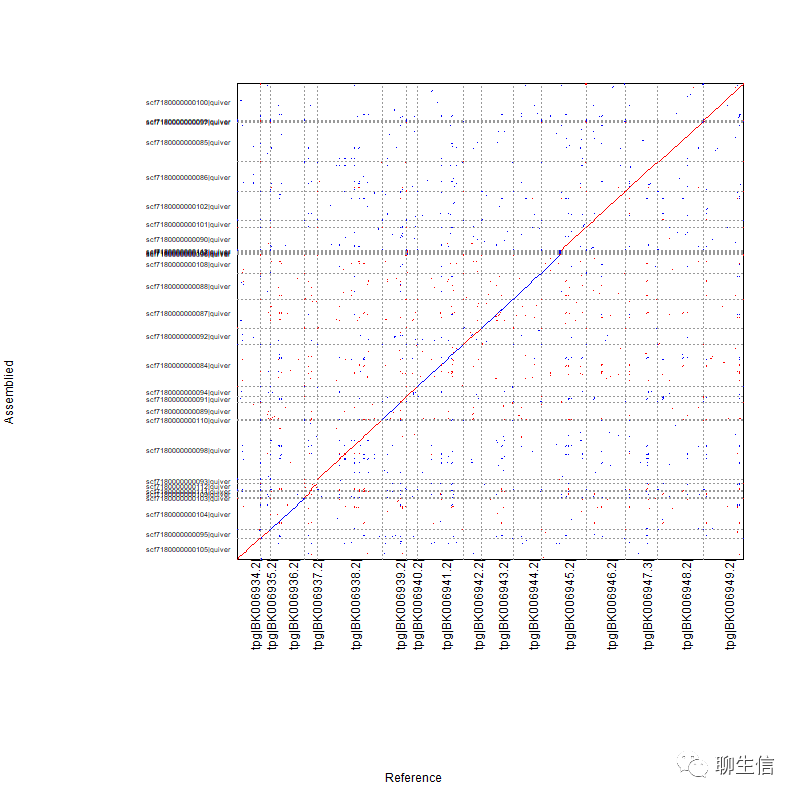
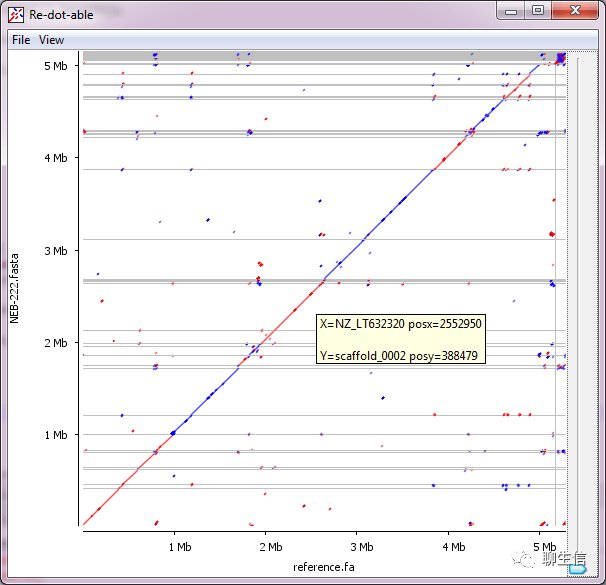
Redottable
https://github.com/s-andrews/redotable
Language: Java
Tags: Dotplot
Platform: Desktop
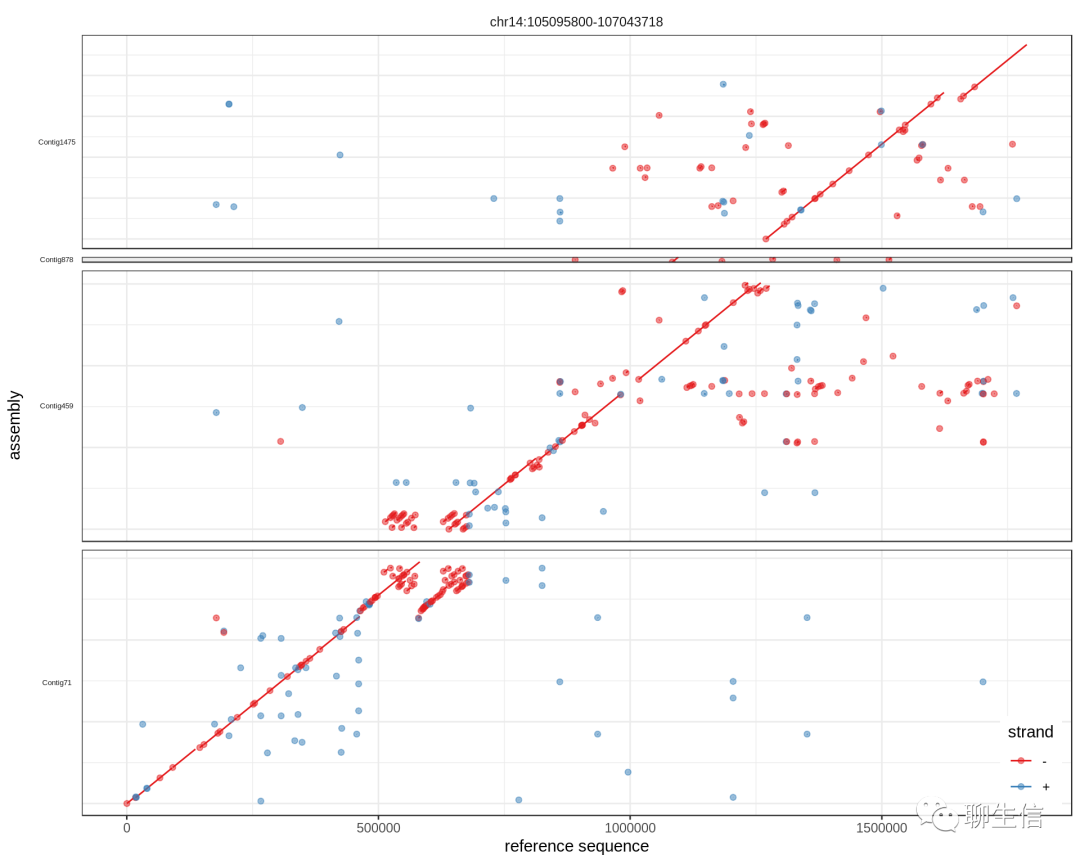
ggplot2 mummerplot
https://jmonlong.github.io/Hippocamplus/2017/09/19/mummerplots-with-ggplot2/
Language: R
Tags: Dotplot
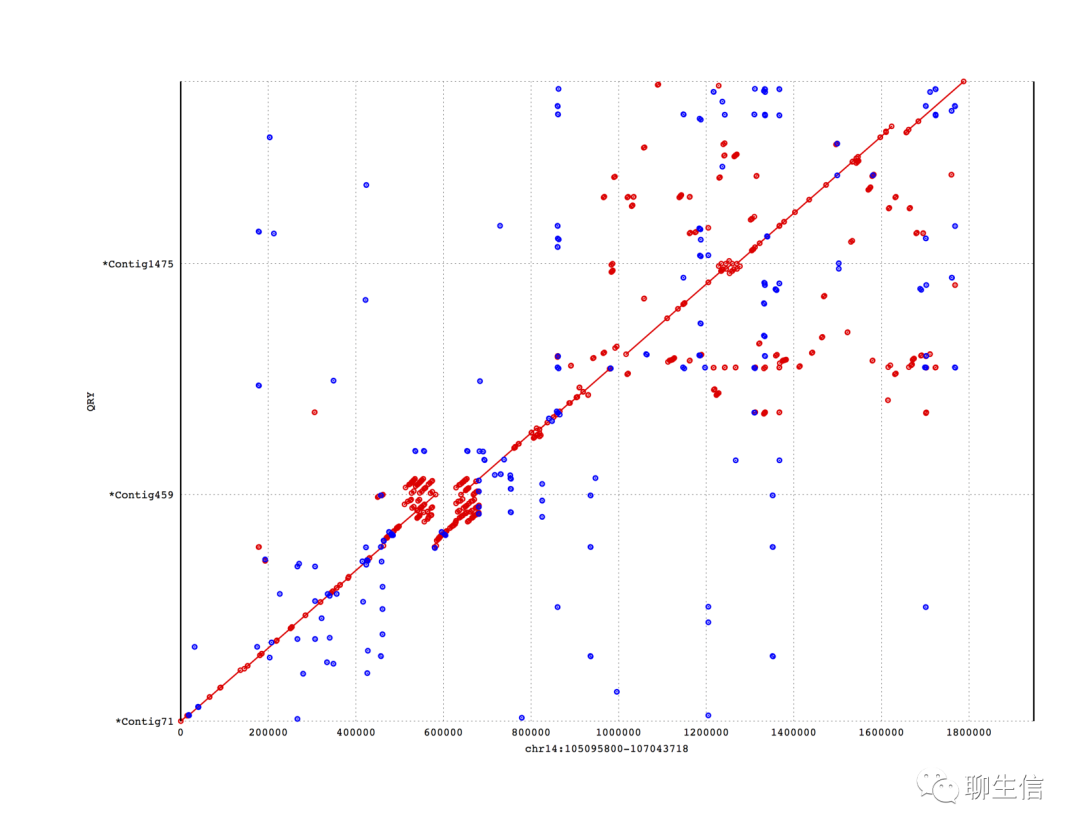
Mummerplot
http://mummer.sourceforge.net/
Language: GNUPlot
Tags: Dotplot, Static
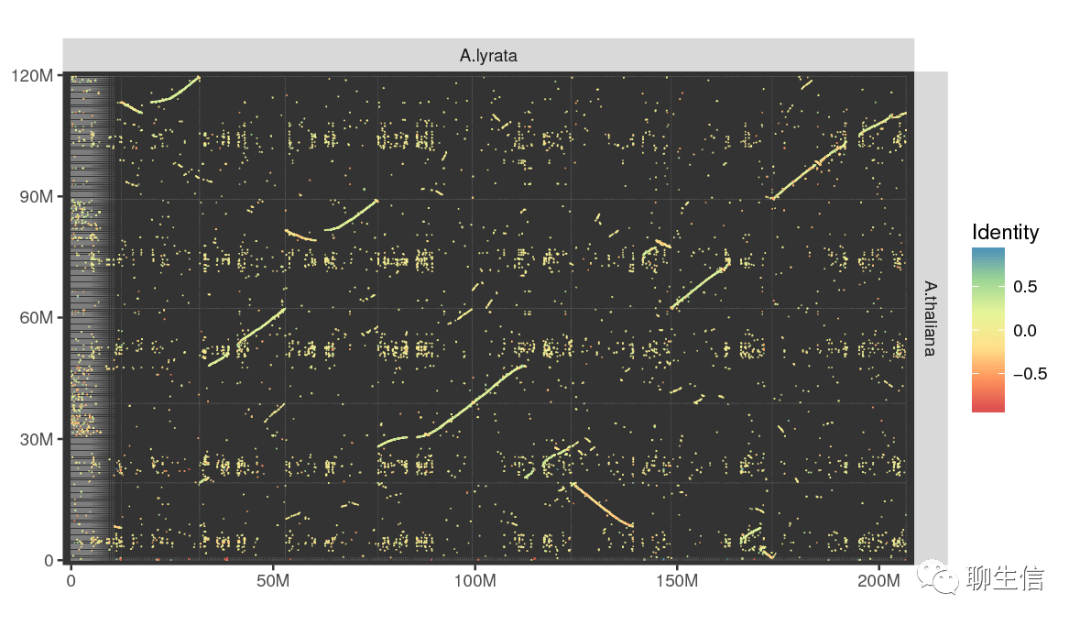
Minidot
https://github.com/thackl/minidot
Language: R
Tags: Dotplot, Static
last-dotplot
http://last.cbrc.jp/
Tags: Dotplot
Platform: CLI, Web-server
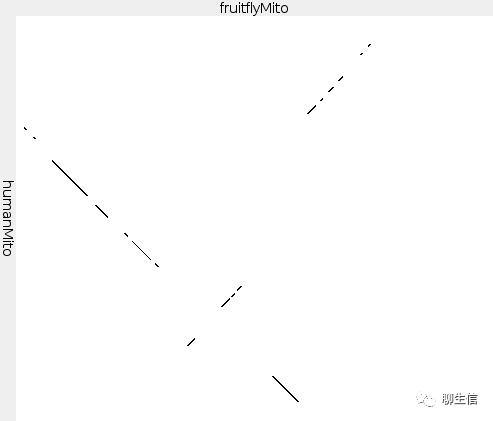
Jdot
https://github.com/LyonsLab/jdot
Language: JS
Tags: Dotplot
Platform: Web

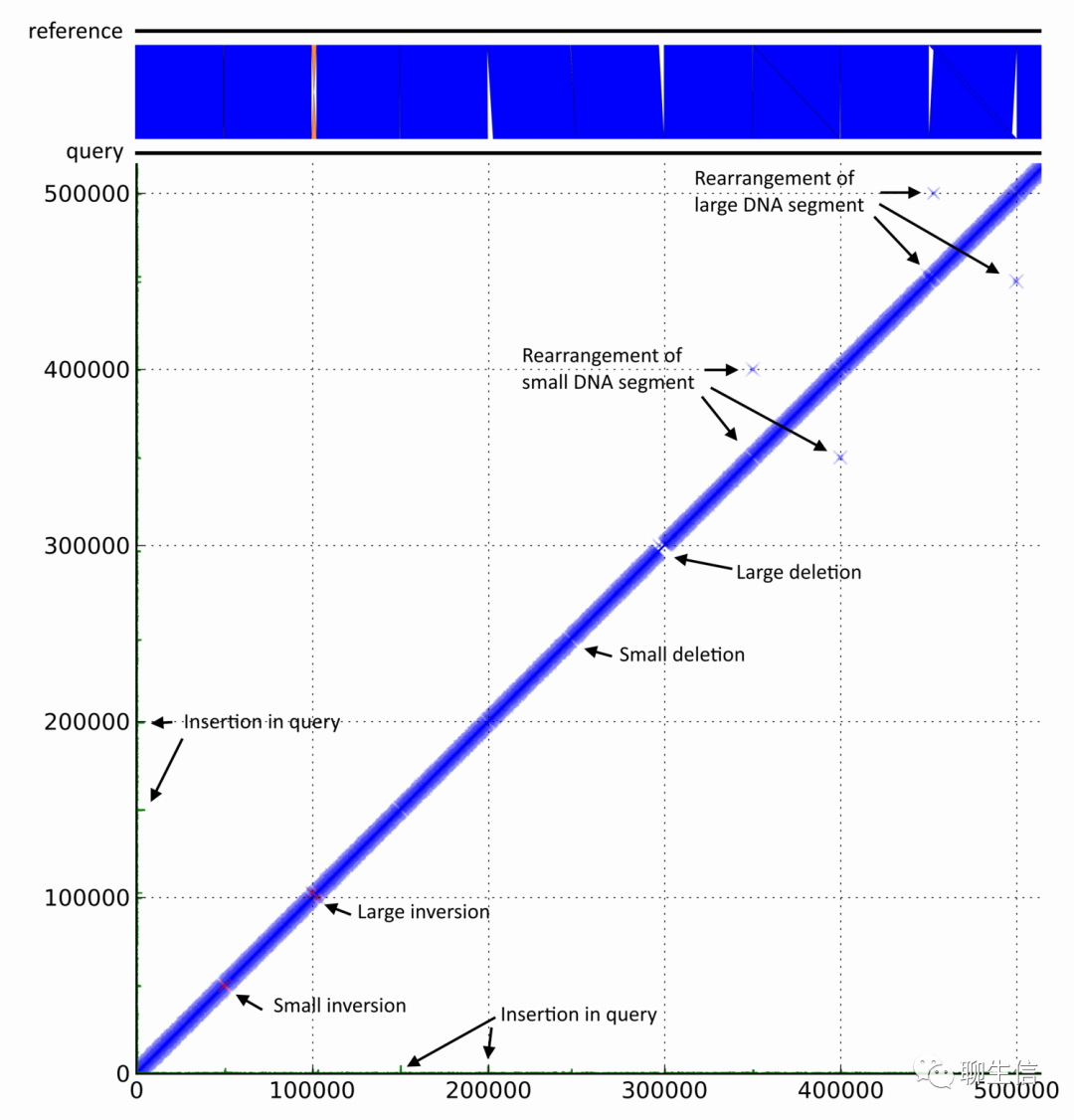
FlexiDot
https://github.com/molbio-dresden/flexidot
Language: Python
Tags: Dotplot
Platform: CLI
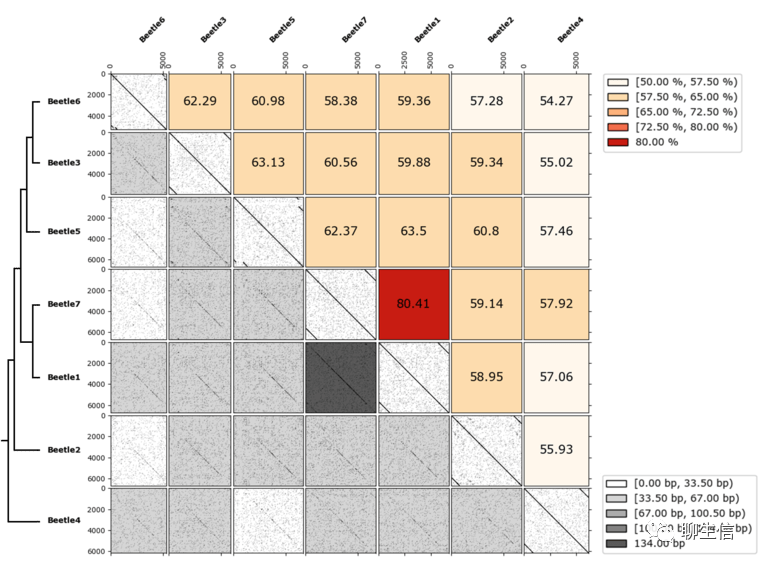
Dottup
https://www.bioinformatics.nl/cgi-bin/emboss/help/dottup
Tags: Dotplot
Note: also seen here http://eichlerlab.gs.washington.edu/publications/chm1-structural-variation/data/GRCh37/heterochromatic_extensions.pdf

dotPlotly
https://github.com/tpoorten/dotPlotly/
Language: R, Shiny
Tags: Dotplot
Platform: Web
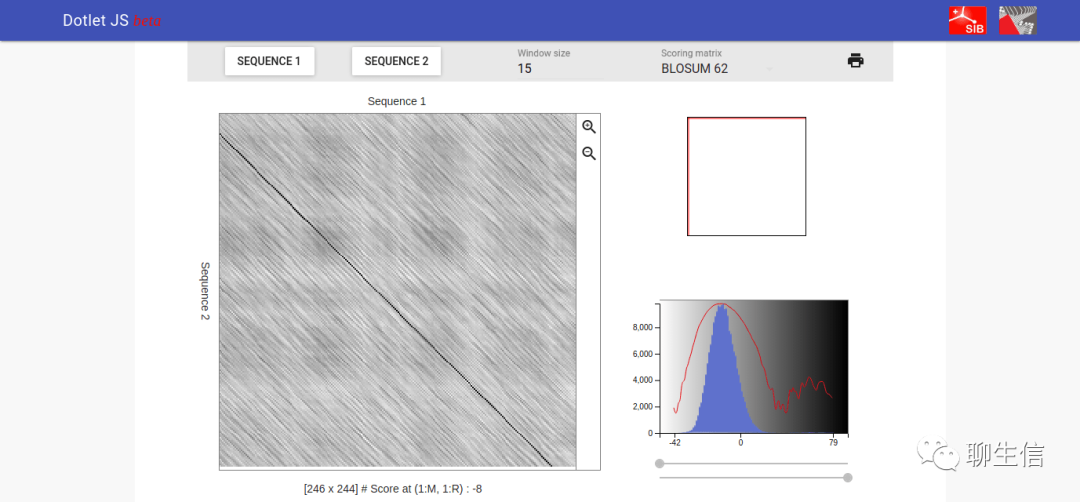
Dotlet
https://dotlet.vital-it.ch/
Language: JS
Tags: Dotplot
Platform: Web
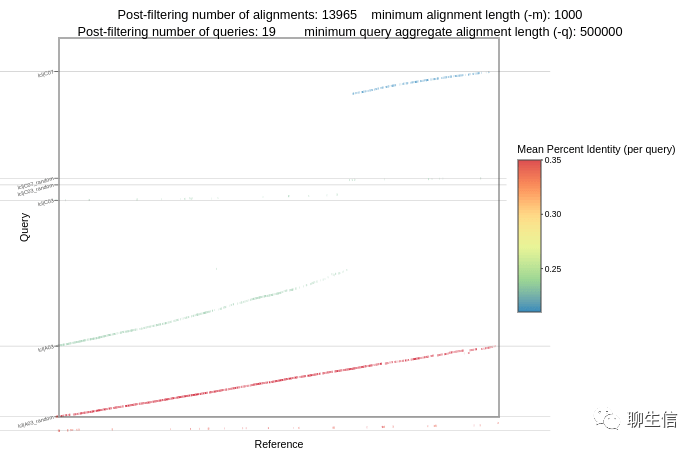
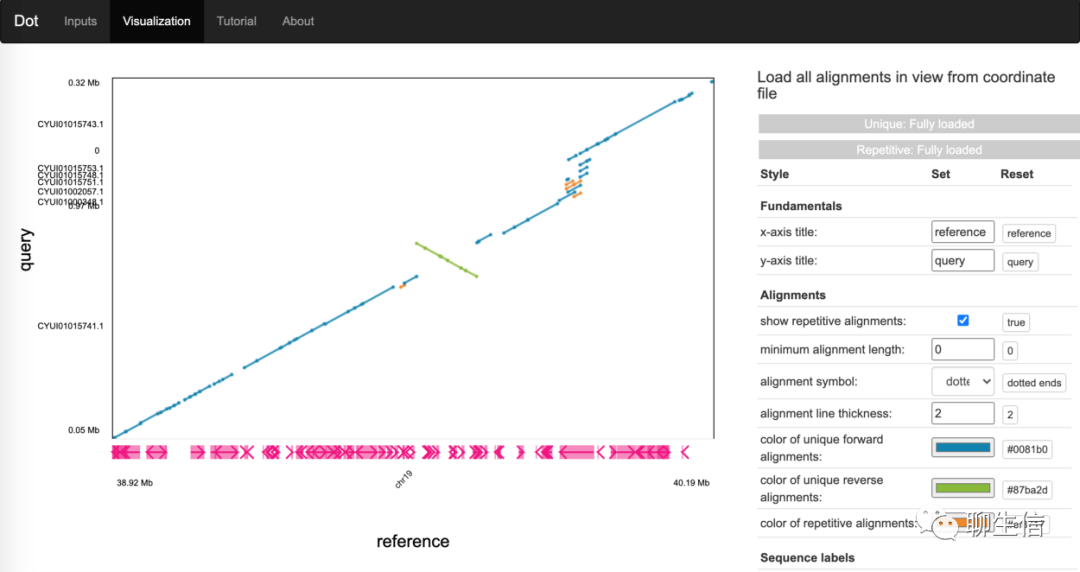
Dot
https://dot.sandbox.bio/
Tags: Dotplot
Github: https://github.com/MariaNattestad/dot
Platform: Web
Discoplot
https://github.com/mjsull/DiscoPlot
Language: Python
Tags: Dotplot, Static
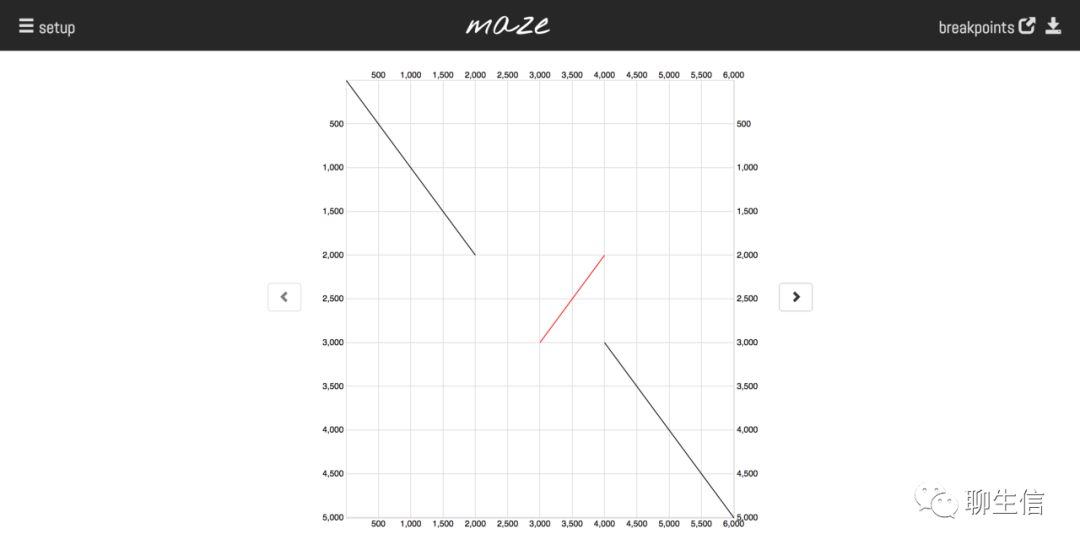
Delly-maze
https://github.com/dellytools/maze
Language: Python
Tags: Dotplot, Static
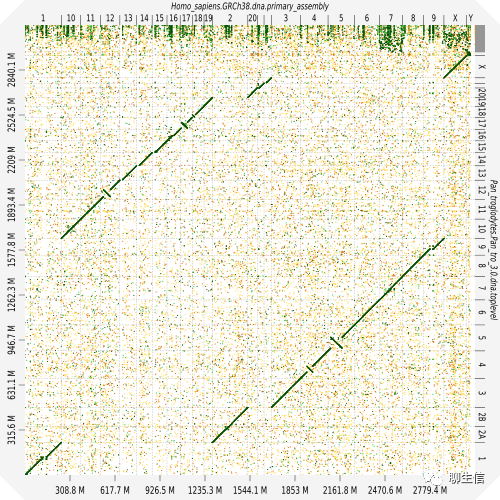
D-GENIES
http://dgenies.toulouse.inra.fr/
Language: JS, Python, D3
Tags: Dotplot
Github: https://github.com/genotoul-bioinfo/dgenies
Platform: Web
cgplot
https://github.com/dfguan/cgplot
Language: Python
Tags: Static, Dotplot
Zenbu
http://fantom.gsc.riken.jp/zenbu/
Tags: General
Platform: Web
WashU epigenomics browser
https://epigenomegateway.wustl.edu/
Language: JS, React
Tags: General, Epigenomics, Hi-C
Platform: Web
Valis browser
https://valis.bio/
Language: JS, WebGL
Tags: General, Quantitative, Epigenomics
Platform: Web
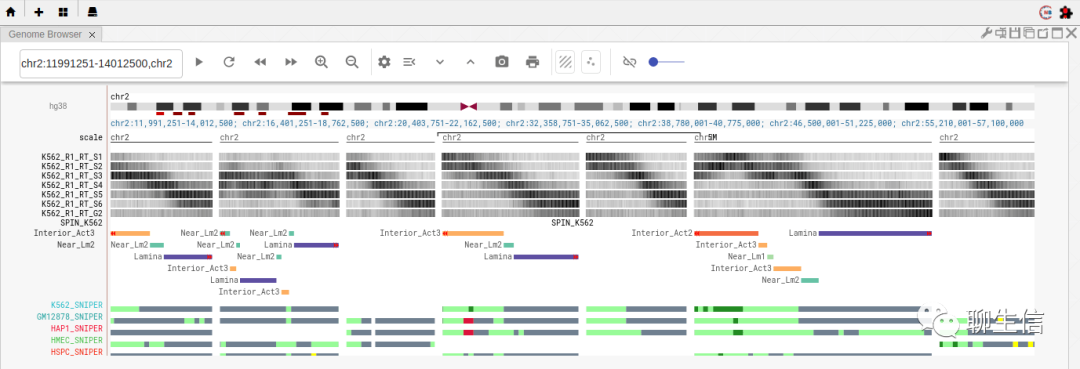
UCSC genome browser
https://genome.ucsc.edu/
Tags: General
Platform: Web, Silo
Trackster
https://galaxyproject.org/learn/visualization/
Tags: General
Platform: Web
Pileup.js
https://github.com/hammerlab/pileup.js/
Language: JS
Tags: General, SV
Platform: Web
Nucleome browser
https://vis.nucleome.org/v1/main.html
Language: JS
Tags: General, Multi
Platform: Web
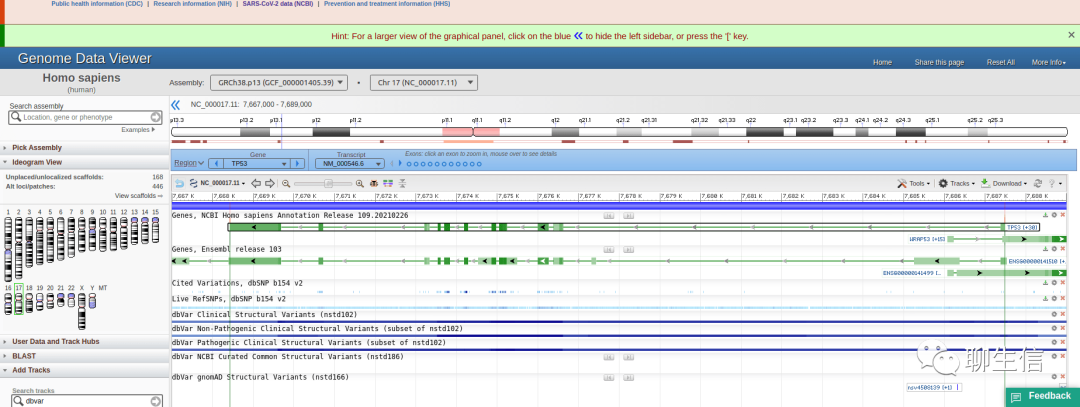
NCBI Genome Data Viewer
https://www.ncbi.nlm.nih.gov/genome/gdv/
Tags: General, SV, Epigenomics, Gene structure, Splicing
Platform: Web, Silo
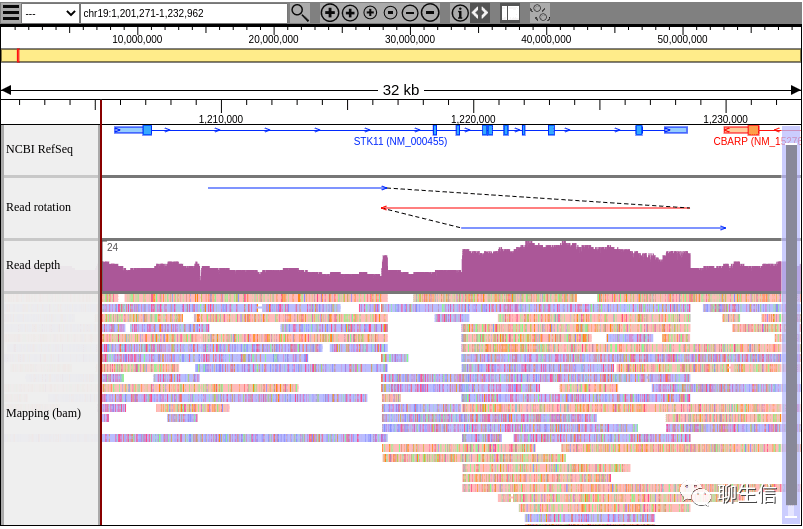
Kero-BROWSE
https://kero.hgc.jp/examples/CLCL/hg38/index.html
Language: JS
Tags: General, SV, Alignment viewer
Platform: Web
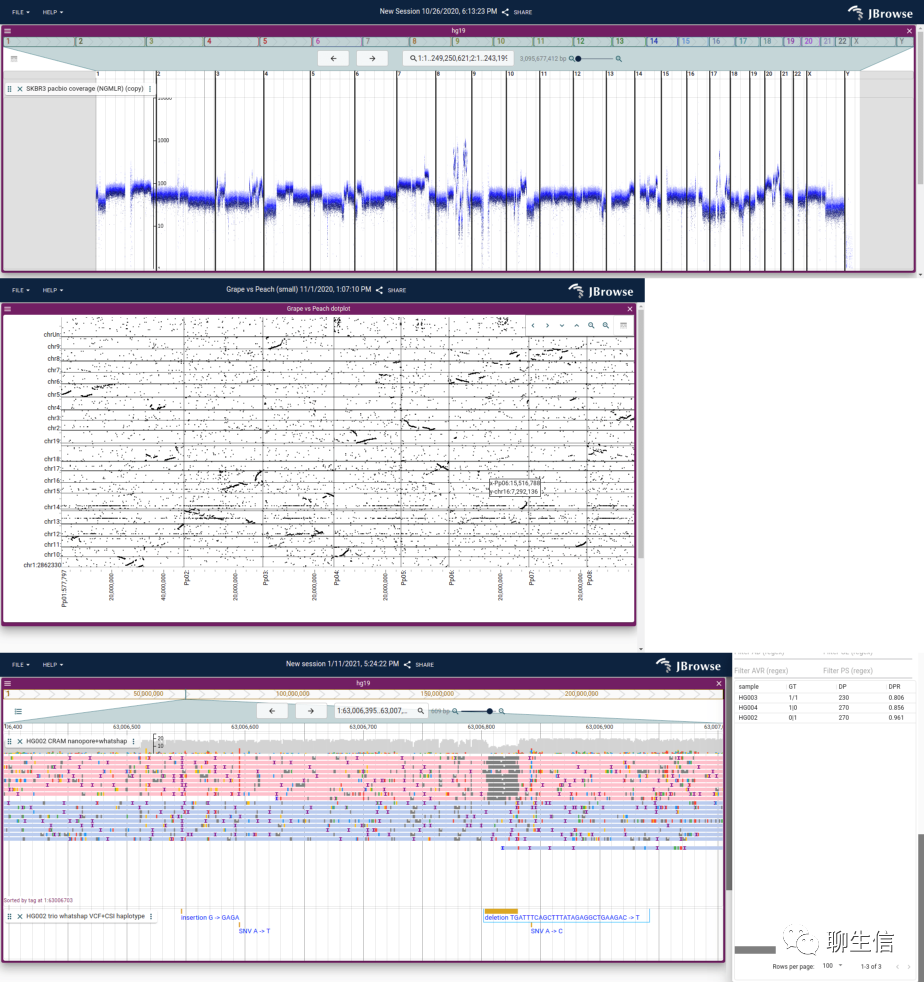
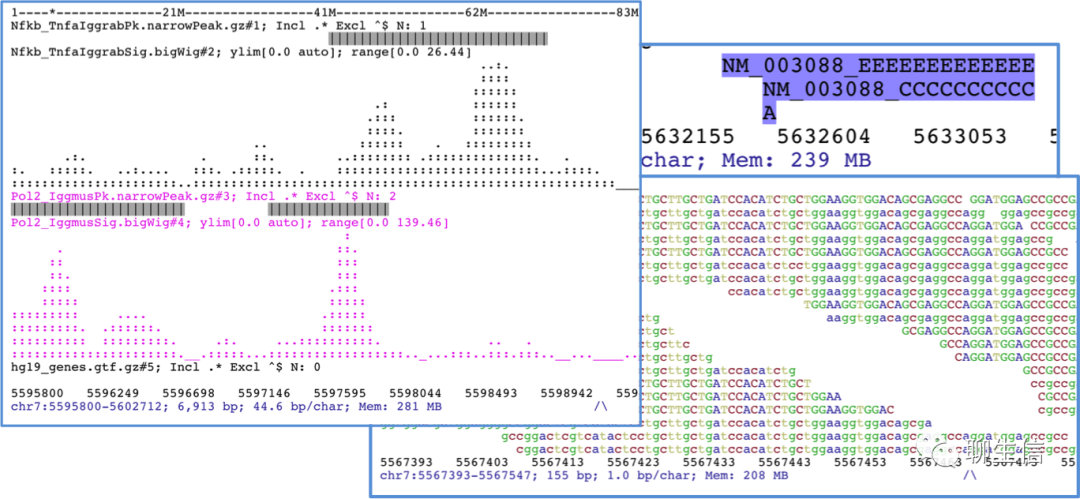
JBrowse 2
http://jbrowse.org/jb2
Language: JS, Typescript, React
Tags: General, SV, Comparative, Dotplot, Circular, Alignment viewer, Quantitative, Hi-C
Note: See gallery for more examples https://jbrowse.org/jb2/gallery
Platform: Web
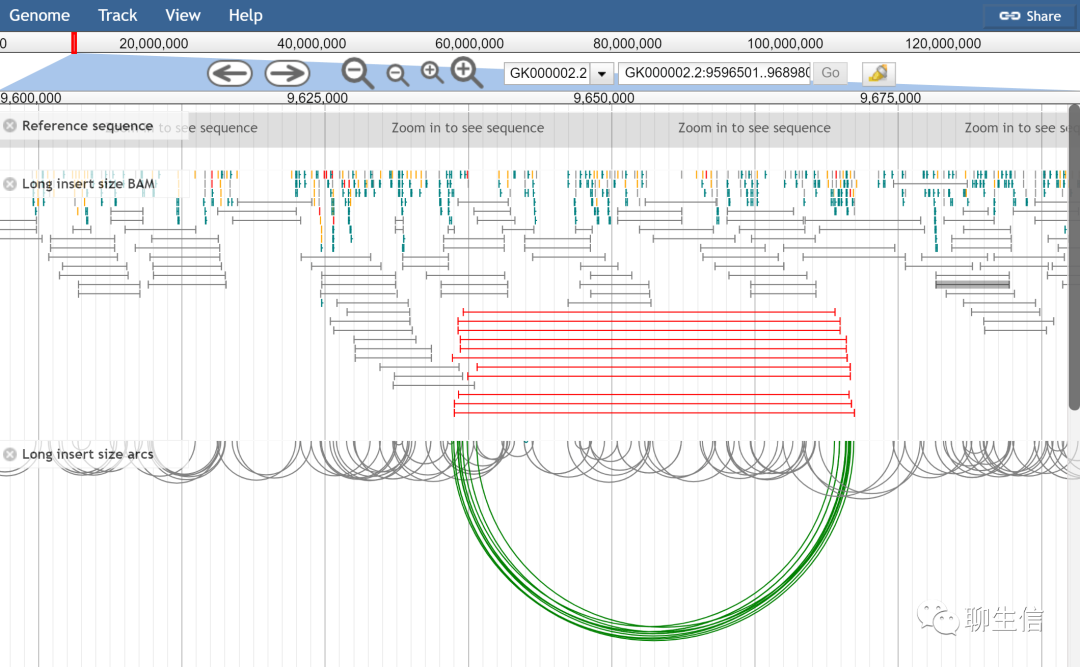
JBrowse
http://jbrowse.org/jbrowse1.html
Language: JS
Tags: General, Alignment viewer, SV, Quantitative, Gene structure
Note: See also JBrowse plugin registry https://gmod.github.io/jbrowse-registry. Runs on the web or as a desktop app using Electron
Platform: Web, Desktop
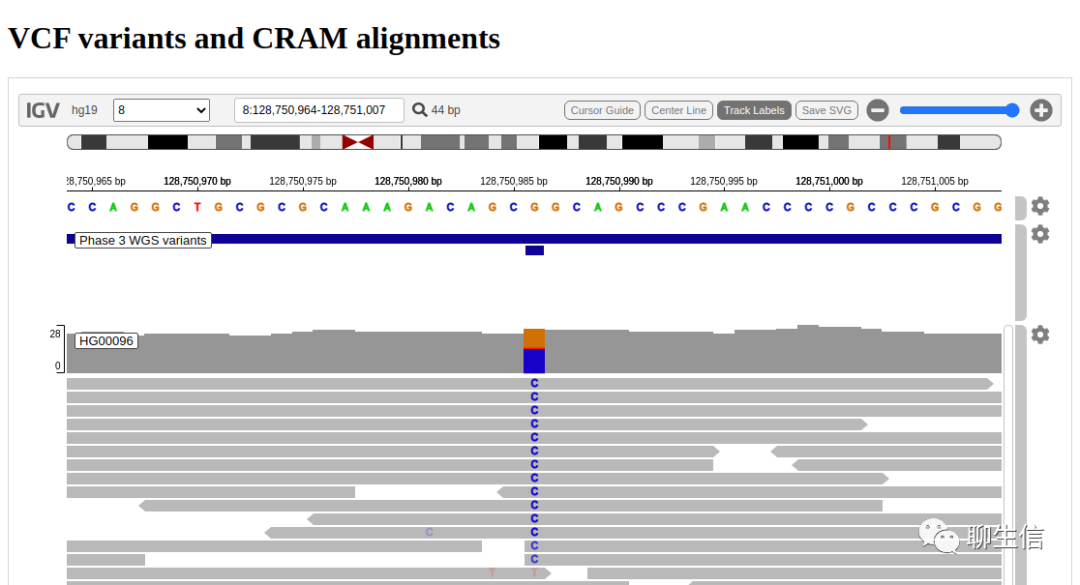
IGV.js
https://github.com/igvteam/igv.js/
Tags: General, Alignment viewer
Platform: Web
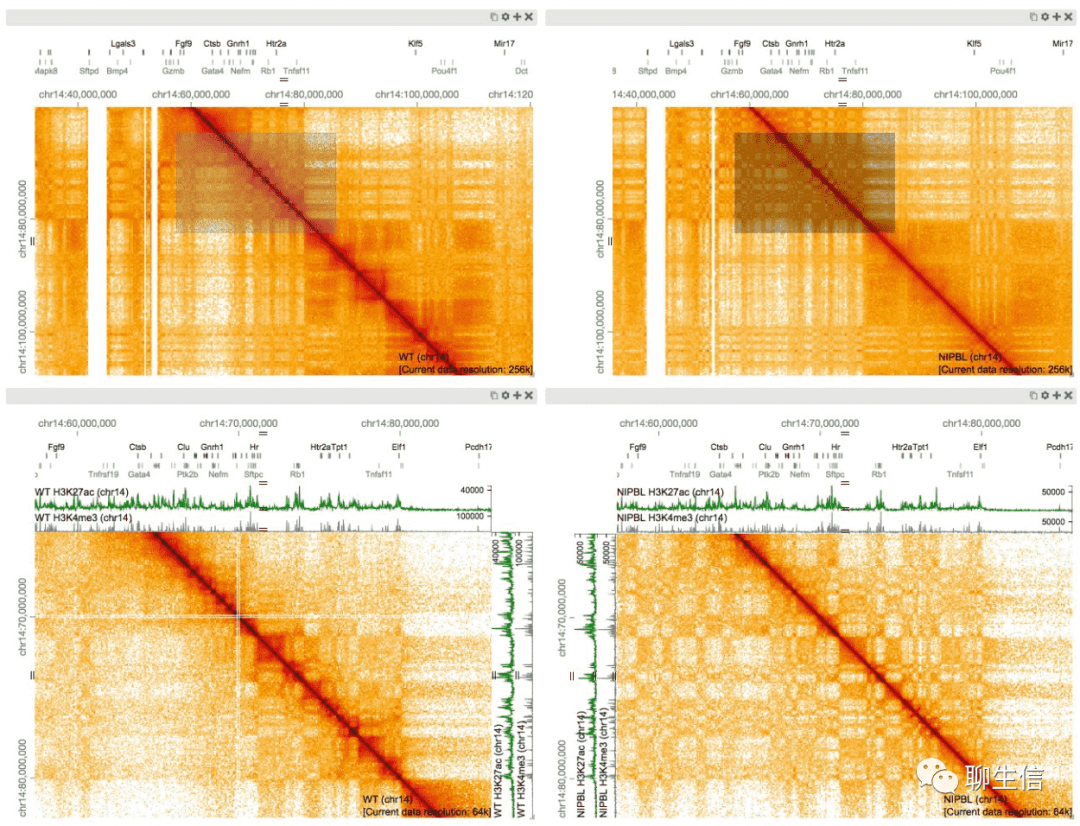
HiGlass
https://higlass.io
Language: JS, WebGL
Tags: General, Epigenomics, Hi-C
Platform: Web
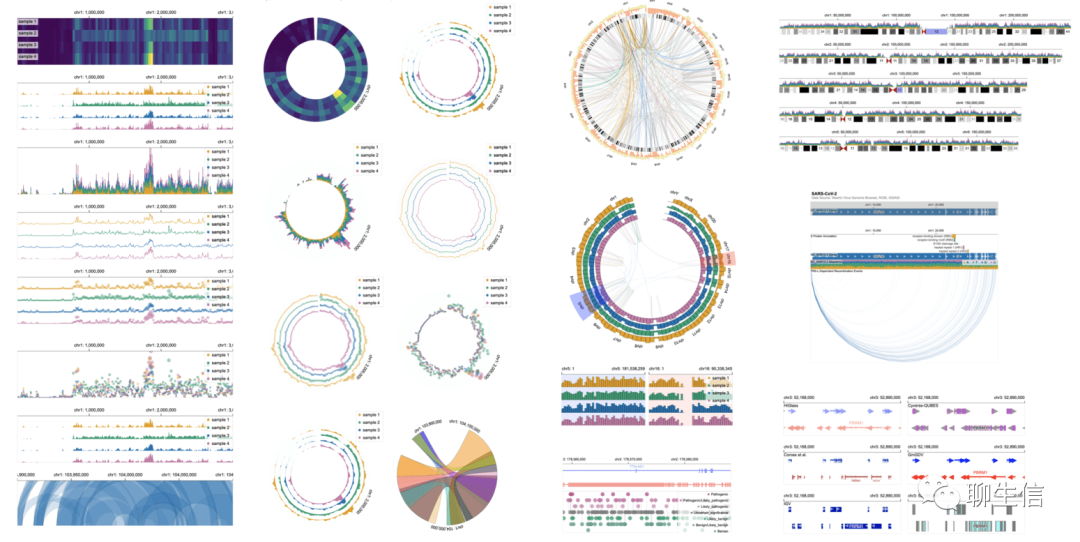
Gosling
https://gosling.js.org/
Language: JS, WebGL, Typescript, React
Tags: General, Epigenomics, Higlass integration, Linear, Circular, Comparative, Quantitative
Platform: Web
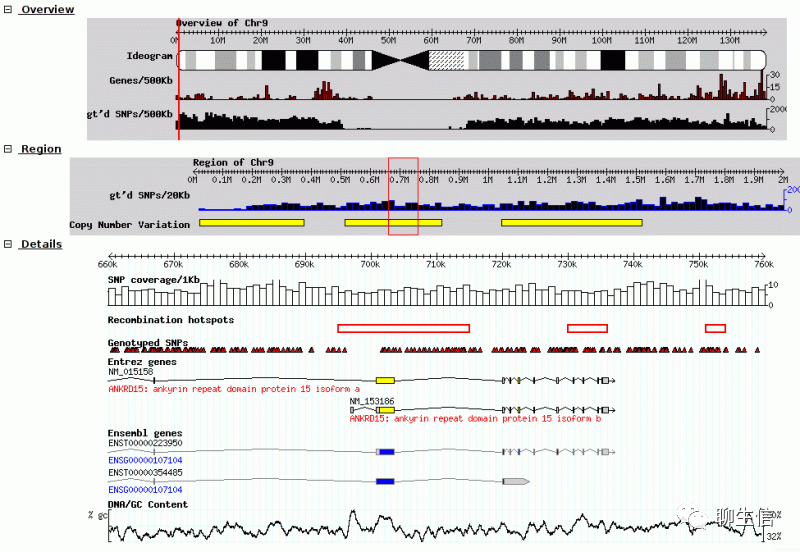
GBrowse+GBrowse 2
http://gmod.org/wiki/GBrowse
Language: Perl
Tags: General
Platform: Web
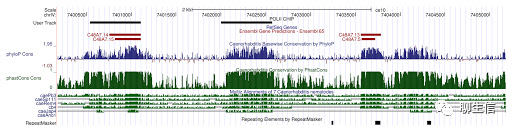
Ensembl genome browser 2020 edition
http://2020.ensembl.org/
Tags: General, Comparative
Platform: Web
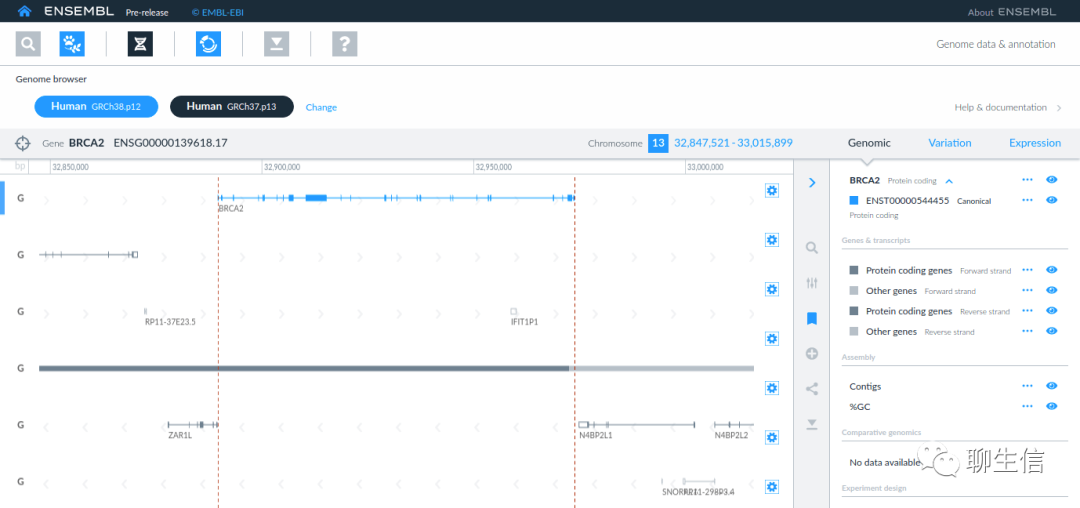
Ensembl genome browser
https://useast.ensembl.org/Homo_sapiens/Location/View?r=17:63992802-64038237
Tags: General, Comparative
Platform: Web, Silo
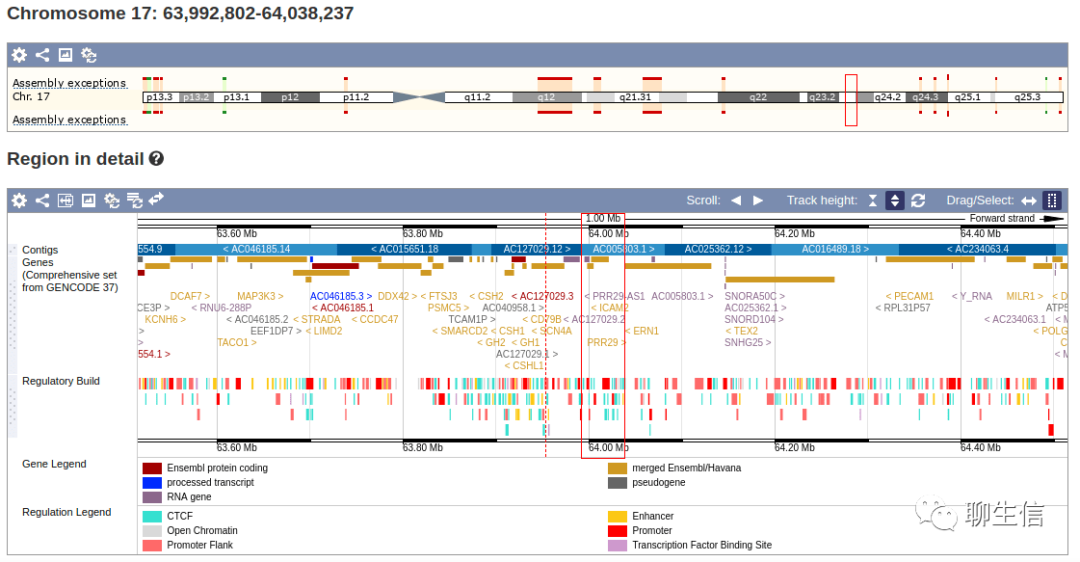
Biodalliance
http://www.biodalliance.org/
Language: JS
Tags: General, Alignment viewer
Platform: Web
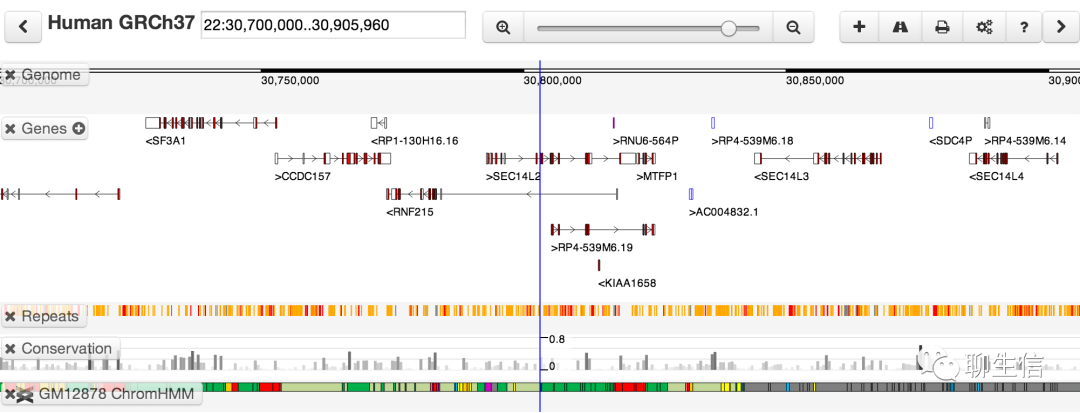
Cylindrical alignment app
https://sourceforge.net/projects/cylindrical-alignment-app/
Alt url https://sourceforge.net/projects/cyl-viewer/
Tags: Exotic
Platform: Desktop
BigTop
https://blog.dnanexus.com/2019-05-21-bigtop-data-visualization/
Tags: Exotic, GWAS
Github: https://github.com/dnanexus/bigtop
Platform: Web

Savant
http://bioinformatics-ca.github.io/savant_genome_browser_lab_2015/
Publication: https://pubmed.ncbi.nlm.nih.gov/22638571/
Language: Java
Tags: General, SV, Alignment viewer
Platform: Desktop
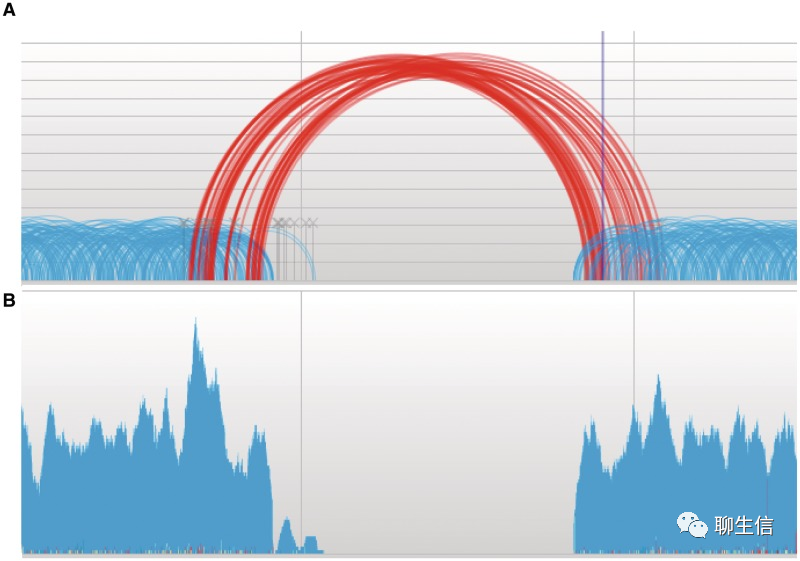
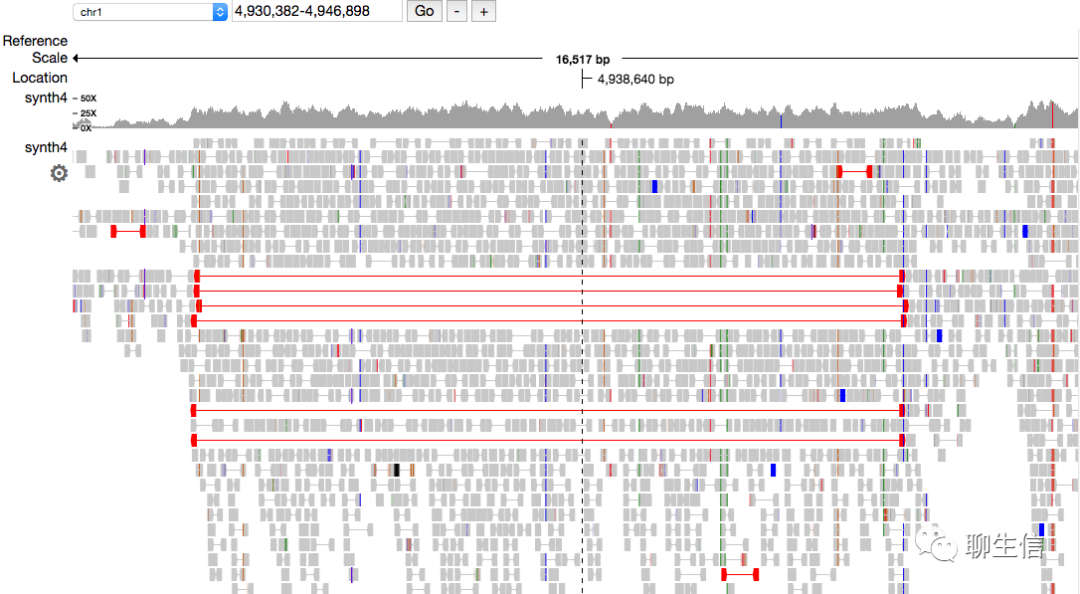
IGV
https://igv.org/
Language: Java
Tags: General, Alignment viewer, SV, Quantitative
Platform: Desktop
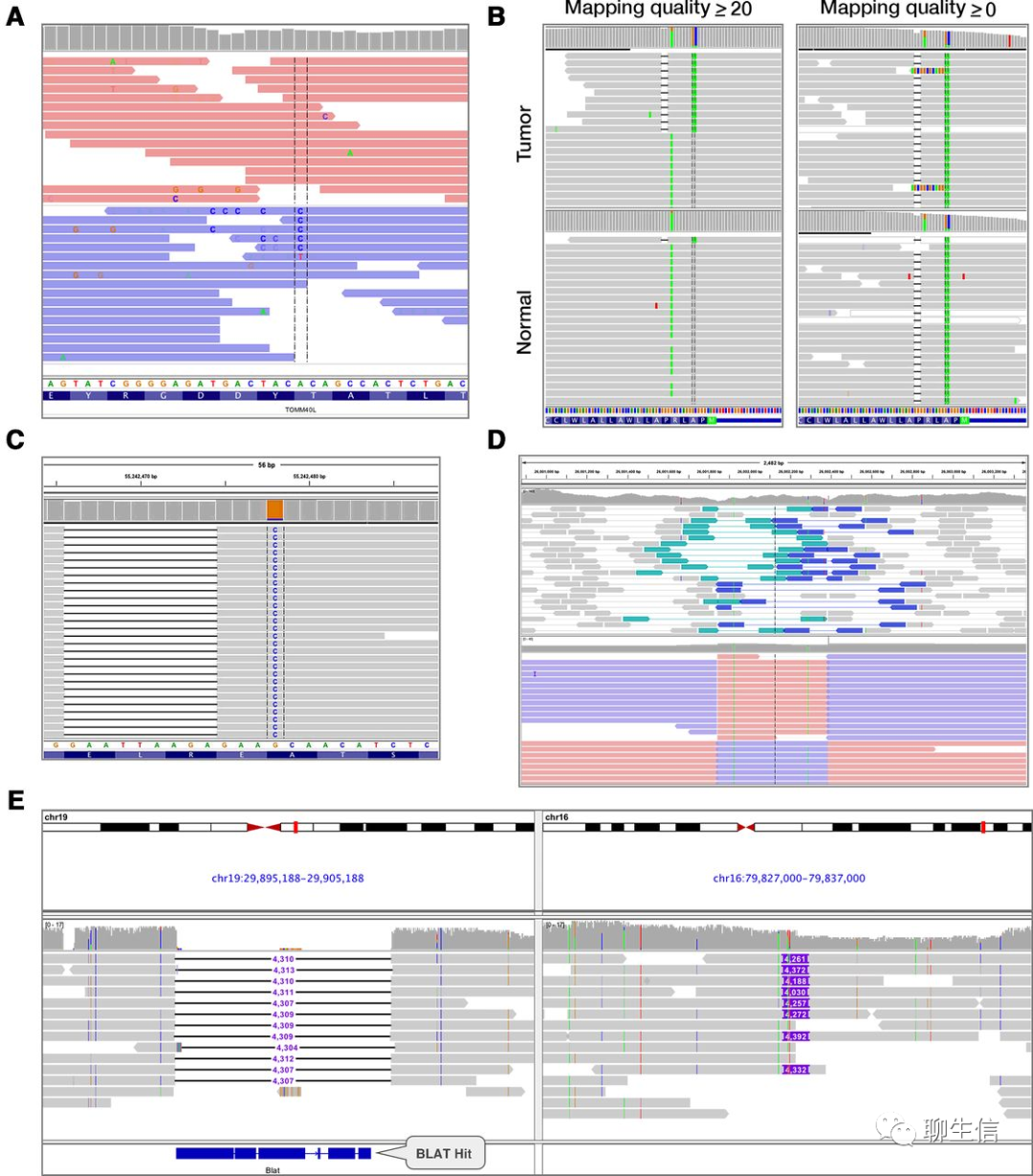
IGB
https://bioviz.org/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4937187/
Language: Java
Tags: General, Alignment viewer
Platform: Desktop
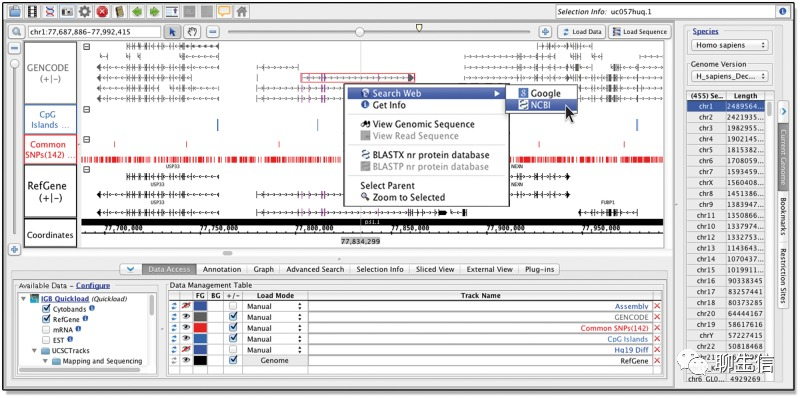
BasePlayer
https://baseplayer.fi/
Publication: https://www.biorxiv.org/content/10.1101/126482v1.full
Language: Java
Tags: General
Platform: Desktop
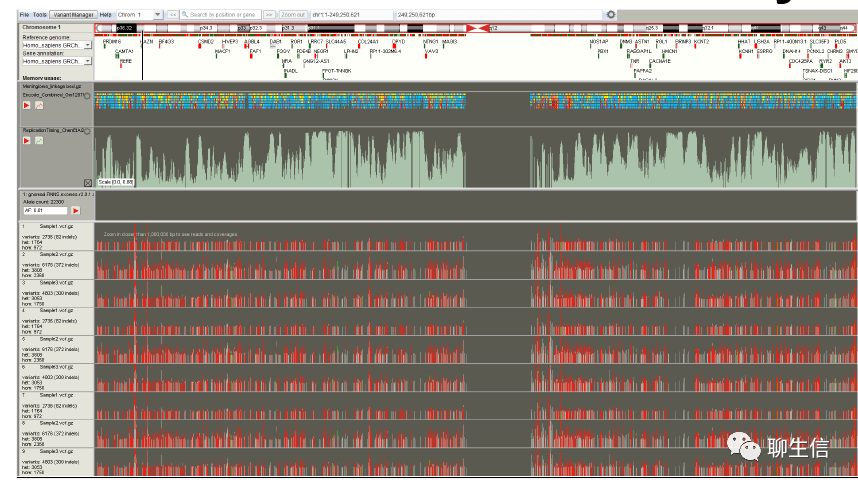
wasm bigwig demo browser
https://shk656461.github.io/index.html
Language: WASM
Tags: Special-purpose, Quantitative
Platform: Web
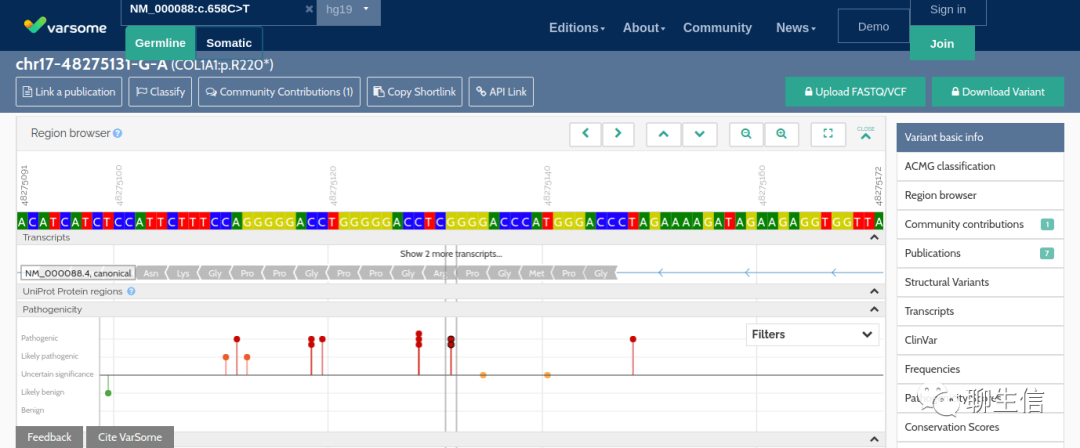
Varsome
https://varsome.com/security-validation/?next=/variant/hg19/NM_000088.3(COL1A1):c.658C%3ET
Tags: Special-purpose, Annotation
Platform: Web, Silo
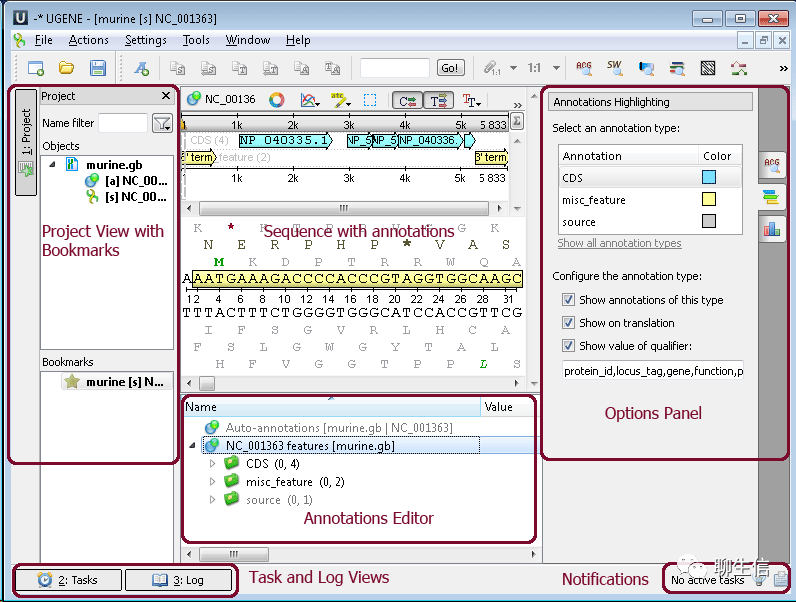
UGENE
http://ugene.net/
Tags: Special-purpose
Platform: Desktop
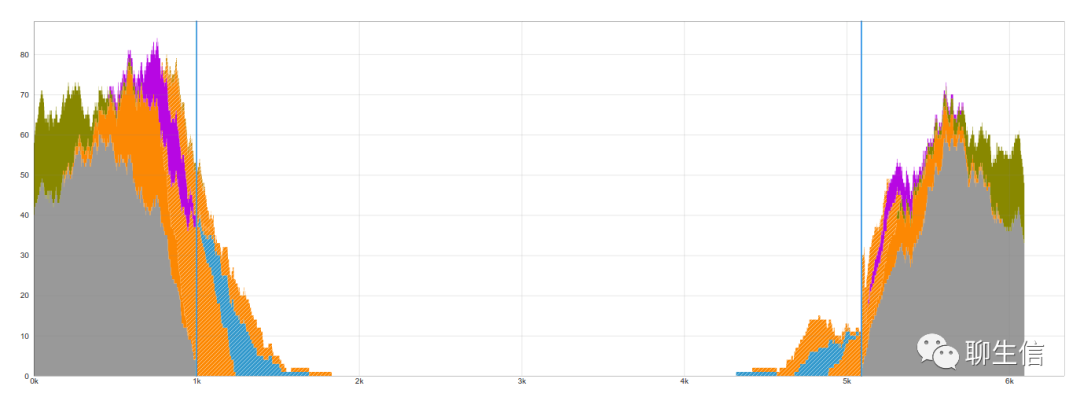
Transposcope
https://academic.oup.com/bioinformatics/article-abstract/36/12/3877/5820971?redirectedFrom=fulltext
Language: JS
Tags: Special-purpose, Transposons
Platform: Web
TE-nest
https://pubmed.ncbi.nlm.nih.gov/18032588
Tags: Special-purpose, Transposons, Deadlink?
Note: download link unknown
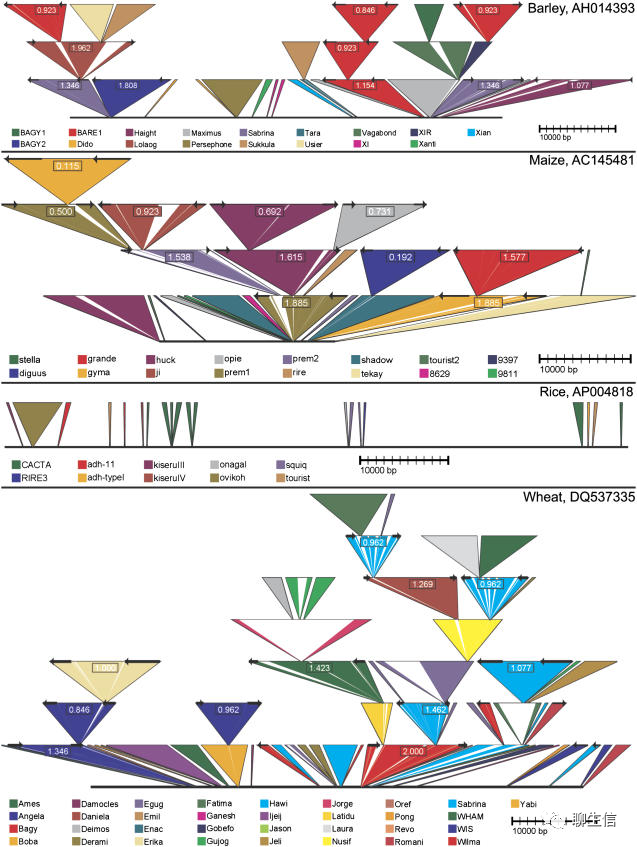
TASUKE
https://ricegenome.dna.affrc.go.jp/
Tags: Special-purpose, Pangenome
Platform: Web
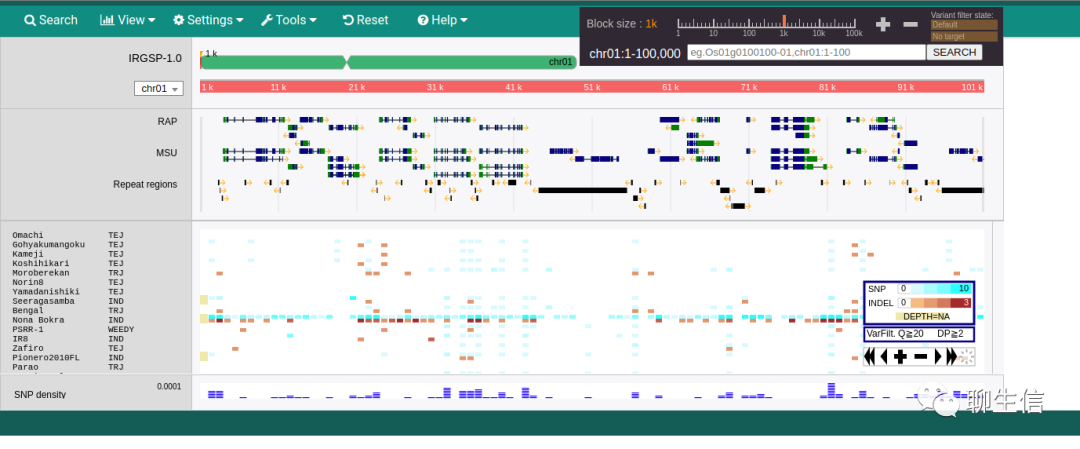
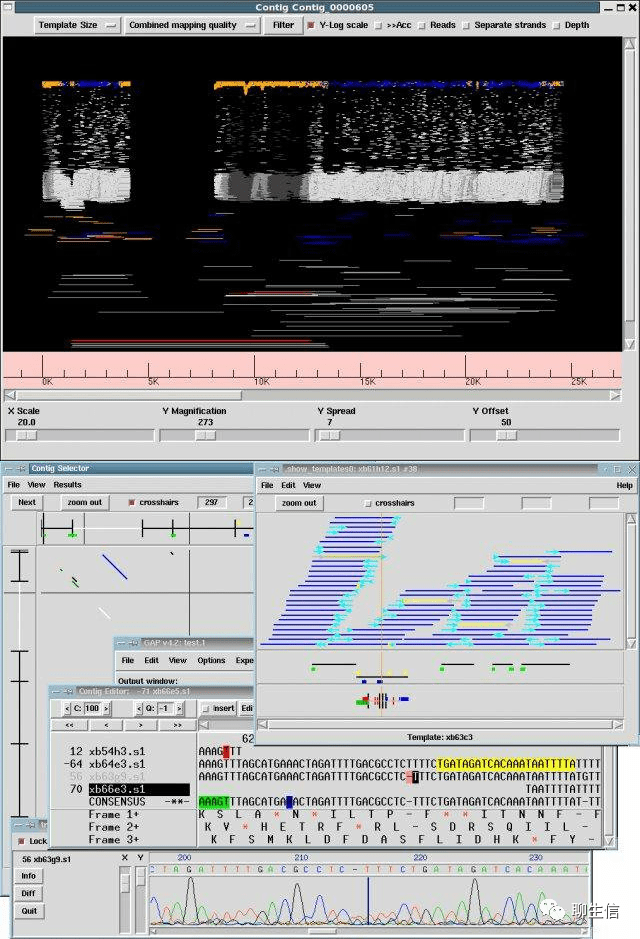
Staden
https://sourceforge.net/projects/staden/
Tags: Special-purpose, Alignment viewer
Platform: Desktop
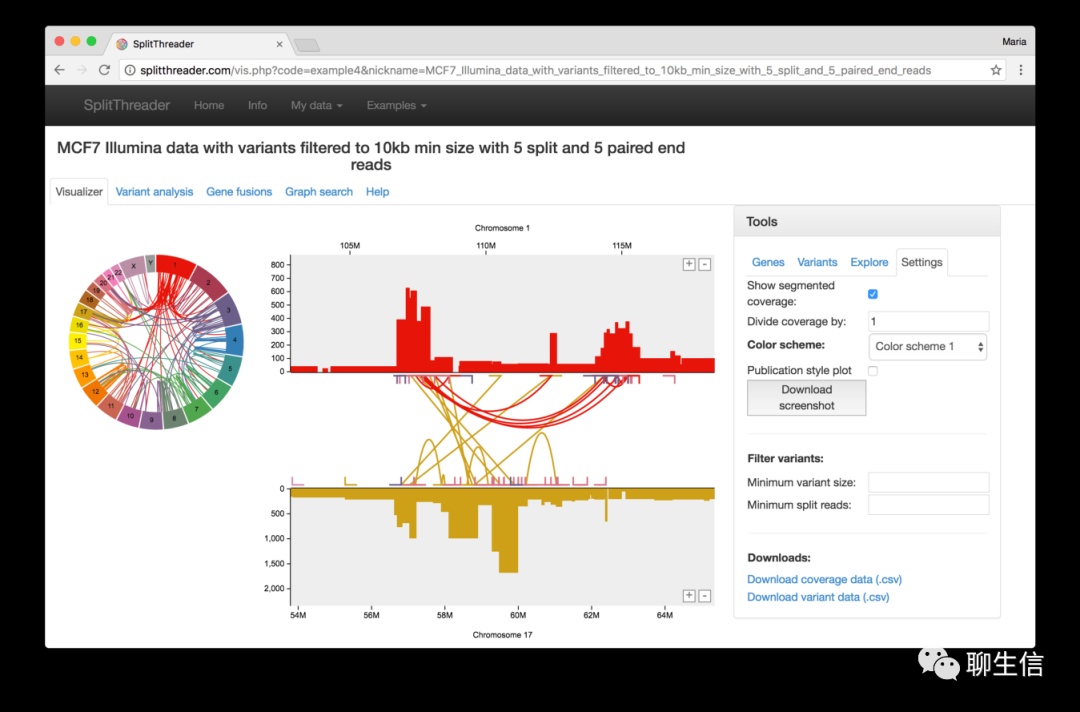
SplitThreader
http://splitthreader.com/
Tags: SV, CNV
Github: https://github.com/MariaNattestad/SplitThreader
Platform: Web
SNPViz
http://www.yeastrc.org/snipviz/4.HTML_Config_retrieve_newick_and_fasta_from_server/snip_viz_HTML_config_with_newick_clustering.html
Tags: MSA
Platform: Web
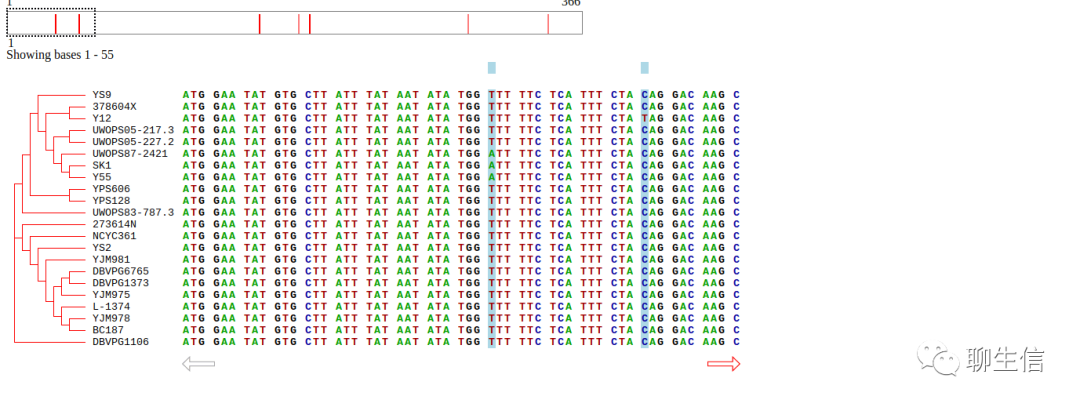
SNPitty
https://bitbucket.org/ccbc/snpitty/src/master/
Publication: https://www.jmdjournal.org/article/S1525-1578(17)30166-6/fulltext
Language: R
Tags: Special-purpose, CNV
Platform: Web
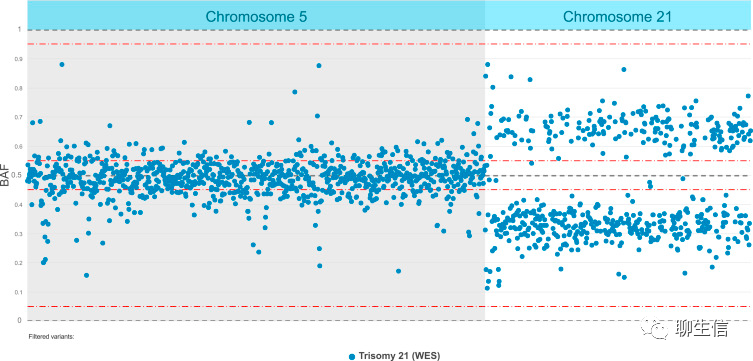
Short read assembly browser
https://rapdb.dna.affrc.go.jp/AssemblyBrowser2/index.html?ref=%22chr01%22&run_id=%22G4toI1%22&start=%22234780%22&end=%22236631%22
Tags: Special-purpose, Alignment viewer
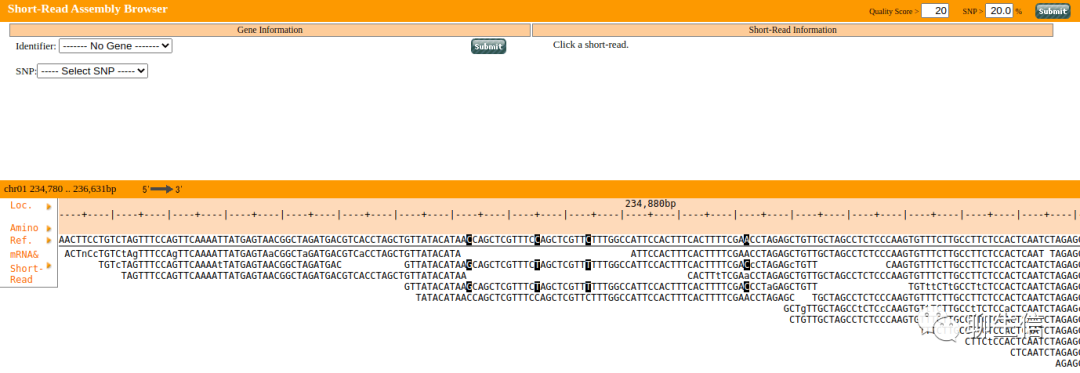
SFARI
https://gene.sfari.org/database/human-gene/
Tags: Special-purpose, CNV
Platform: Web, Silo
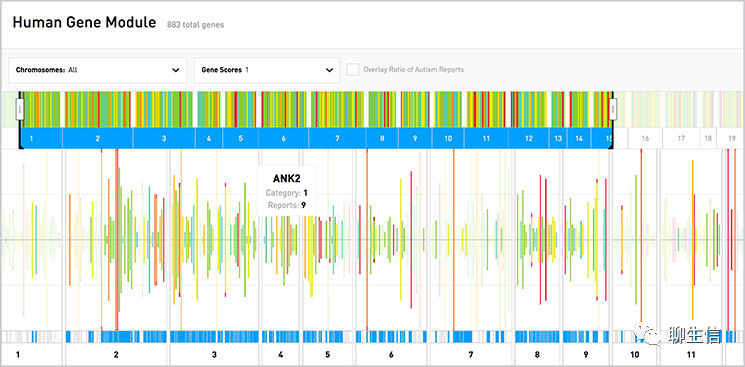
SEQing
https://github.com/malewins/SEQing
Language: Python
Tags: Special-purpose
Platform: Web
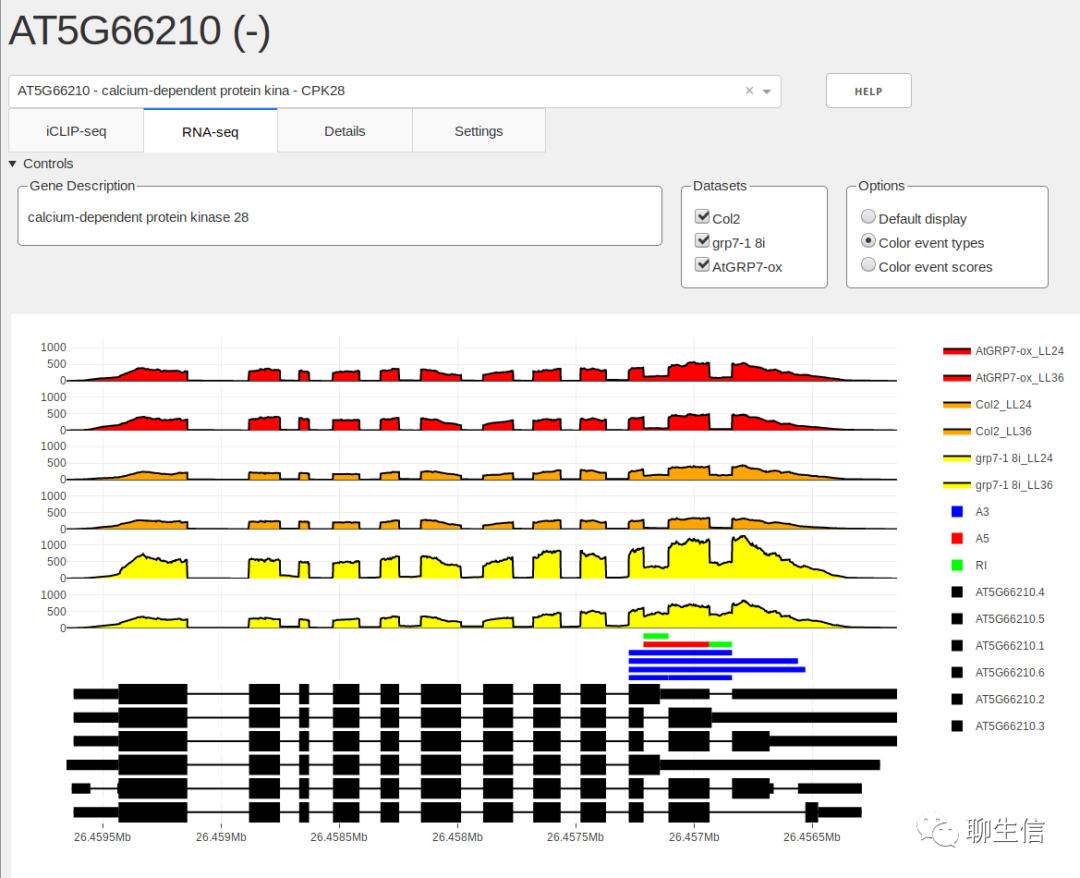
SeqCover
https://brentp.github.io/seqcover/#gene=AIFM1
Tags: Special-purpose, Coverage
Platform: Web
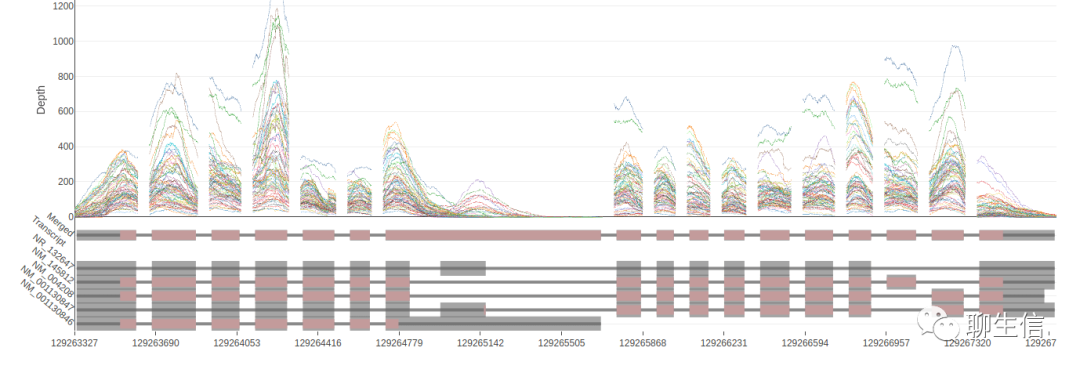
NGB
https://github.com/epam/NGB
Language: Java
Tags: Special-purpose, Alignment viewer
Platform: Web
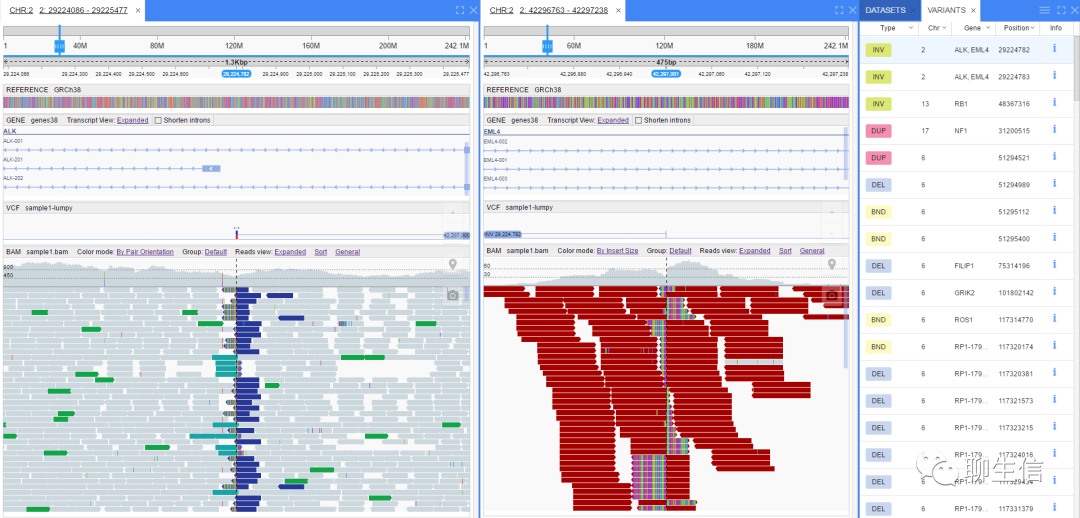
MapView
https://sites.google.com/site/wjwdavy/
Publication: https://academic.oup.com/bioinformatics/article/25/12/1554/193807
Tags: Special-purpose, Alignment viewer
Note: links in original paper are dead, but URL provided here works
Platform: Desktop
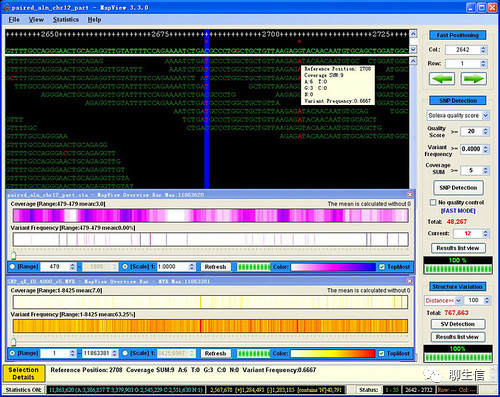
Panoptes
https://www.malariagen.net/apps/ag1000g/phase1-AR3/index.html
Tags: Special-purpose, Population
Github: https://github.com/cggh/panoptes
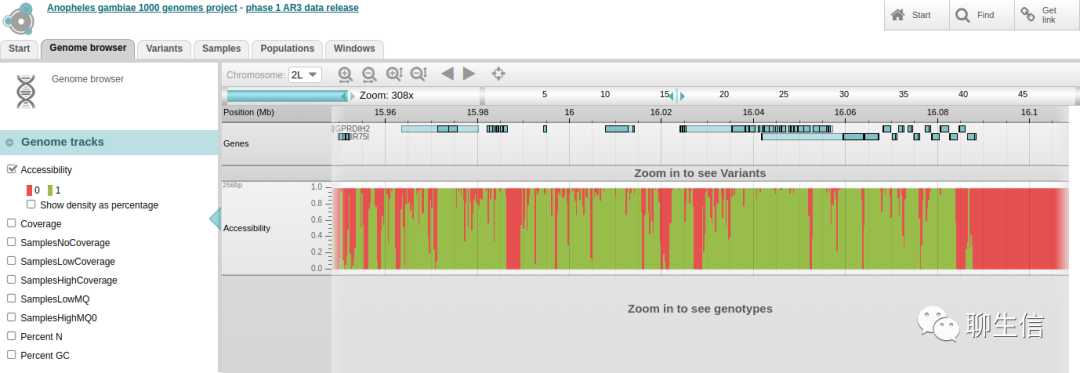
LookSeq
https://www.sanger.ac.uk/tool/lookseq/
Tags: Alignment viewer
Platform: Web
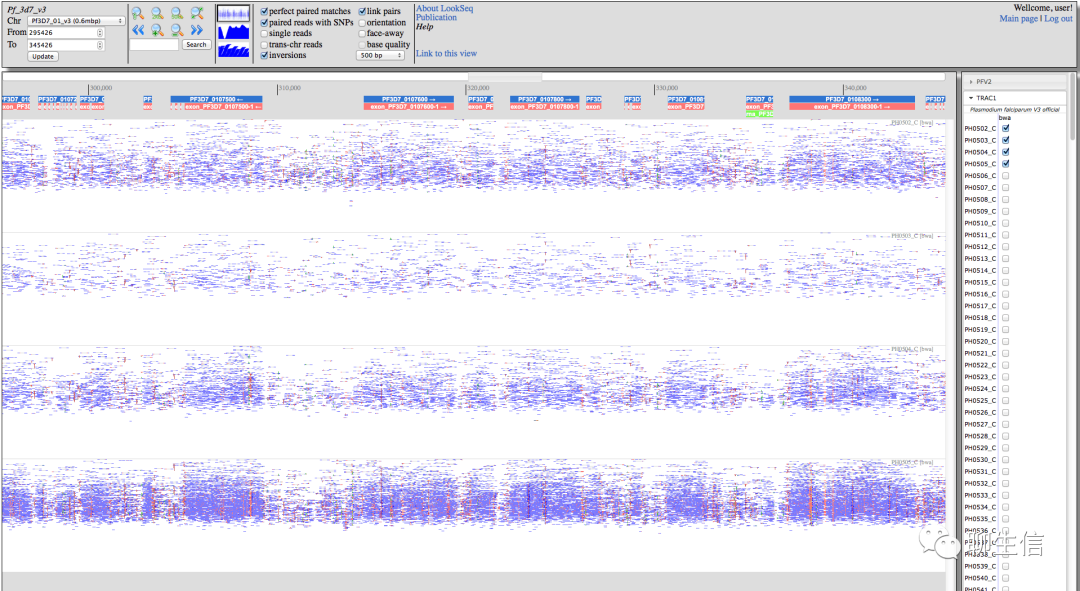
Locuszoom
http://locuszoom.org/
Tags: GWAS, Population
Note: Also see locuszoom.js http://locuszoom.org/locuszoomjs.php
Platform: Web
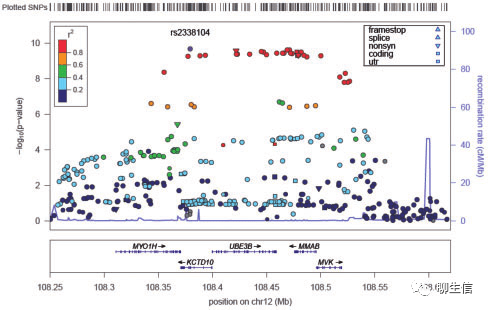
IslandPlot
https://bl.ocks.org/lairdm/c6c235dbfa6e6ee61565
Language: JS
Tags: Special-purpose
Note: D3, SVG
Platform: Web
Introgression browser
https://git.wageningenur.nl/aflit001/ibrowser
Publication: https://onlinelibrary.wiley.com/doi/pdf/10.1111/tpj.12800
Language: Python
Tags: SV, Introgression
Platform: Web
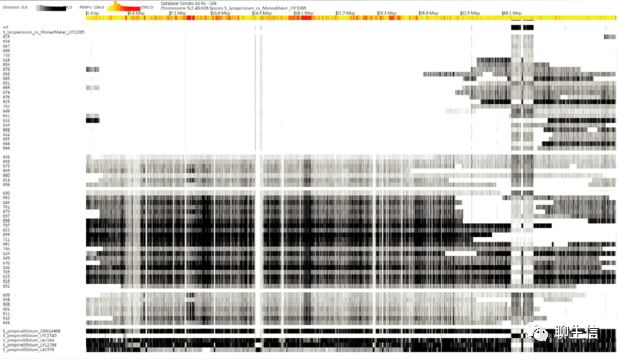
InGAP-SV
http://ingap.sourceforge.net/
Tags: SV, Alignment viewer
Platform: Desktop
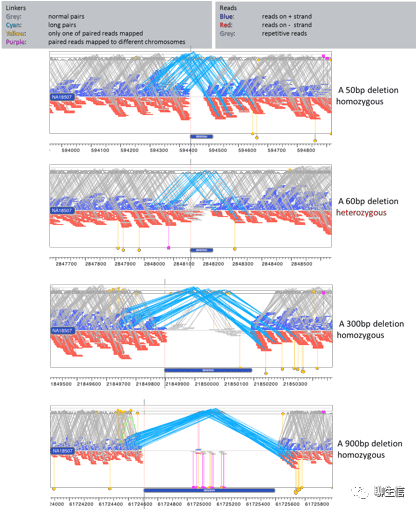
Icarus
http://quast.sourceforge.net/icarus.html
Tags: Assembly QC
Platform: Web
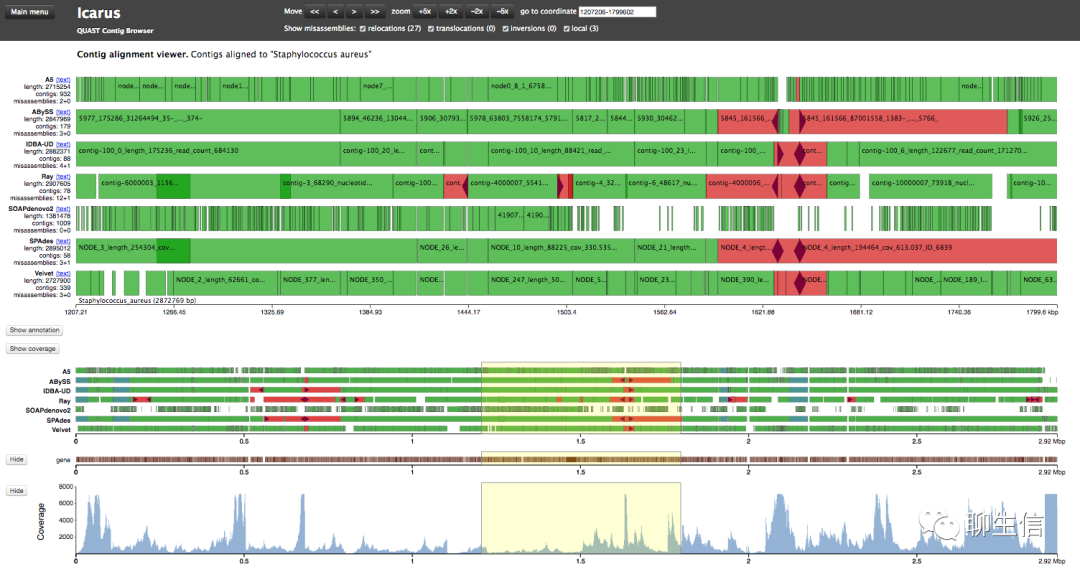
Human genome dating
https://human.genome.dating/region/chr1_13250000
Tags: Population, GWAS
Note: made with vega/d3
Platform: Web
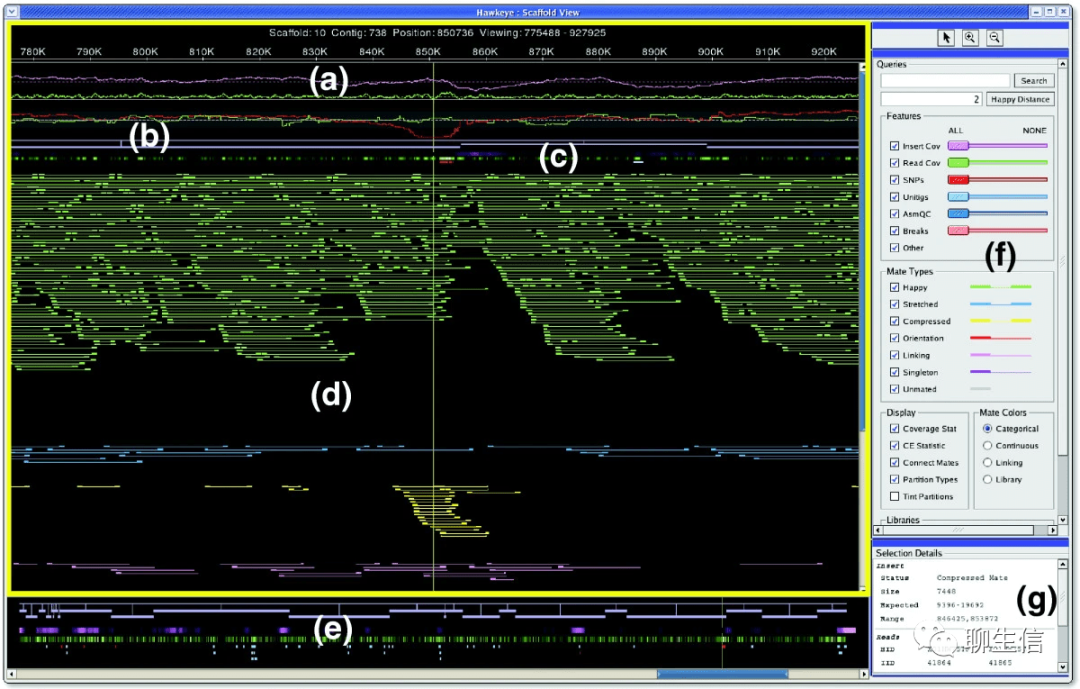
Hawkeye
Publication: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2007-8-3-r34 (2007)
Tags: Special-purpose, Alignment viewer, Assembly QC
Platform: Desktop
GWAS catalog browser
https://www.ebi.ac.uk/gwas/variants/rs1558902
Tags: Population, LD, GWAS
Platform: Web
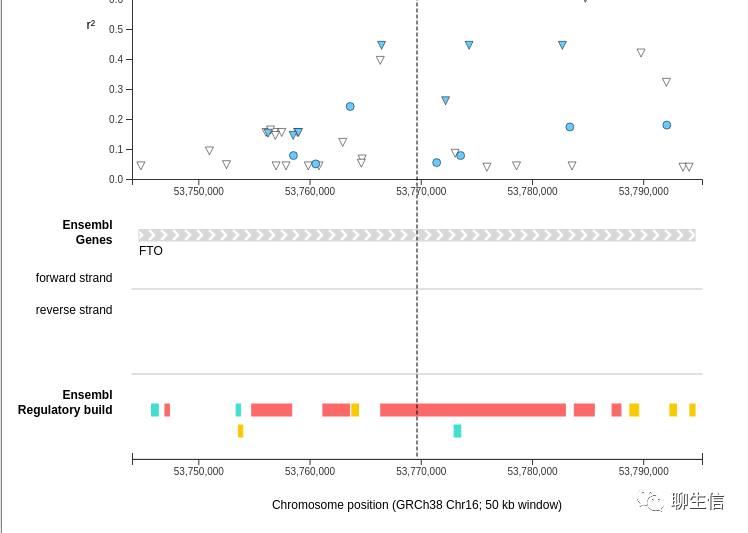
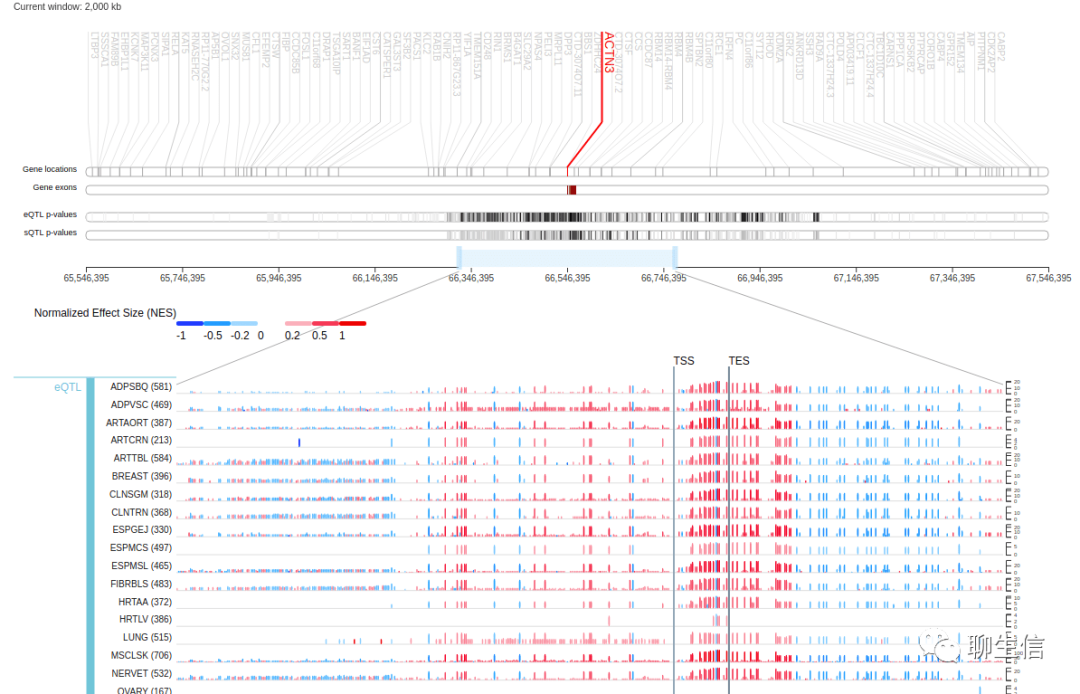
GTEX Locus browser
https://gtexportal.org/home/locusBrowserPage/ACTN3
Tags: Expression, Linear, LD
Platform: Web
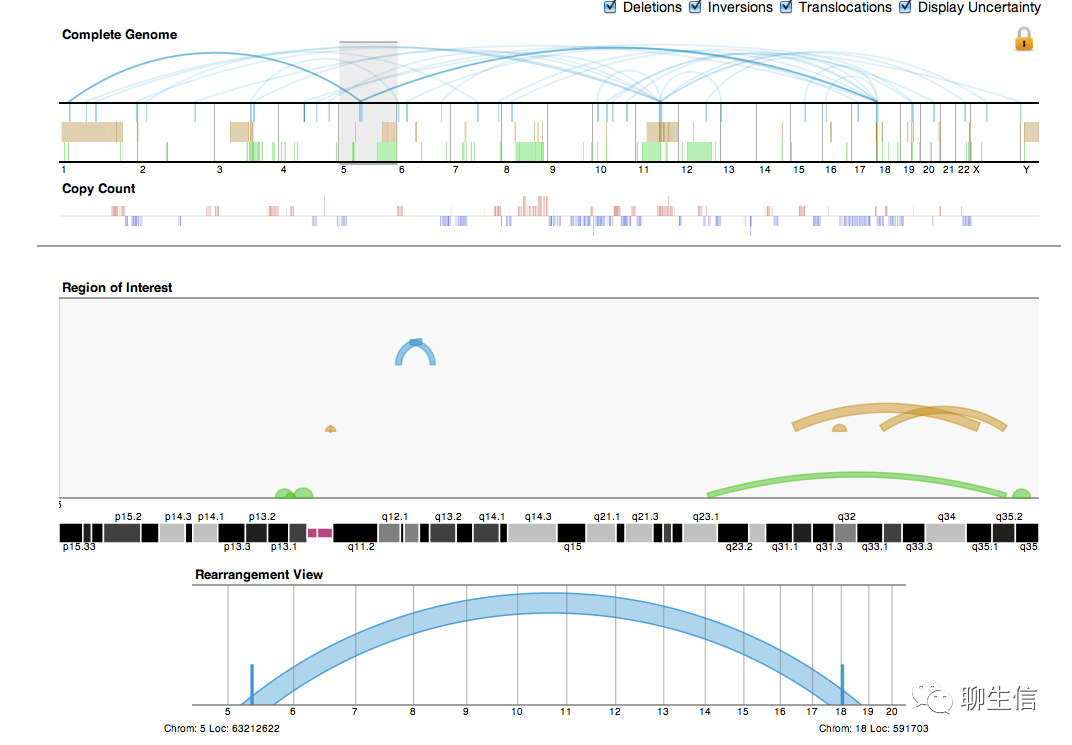
Gremlin
http://compbio.cs.brown.edu/projects/gremlin/
Tags: SV, Linear
Platform: Web
Gnomad browser
https://github.com/macarthur-lab/gnomadjs
Tags: Special-purpose, Linear, Population
Platform: Web, Silo
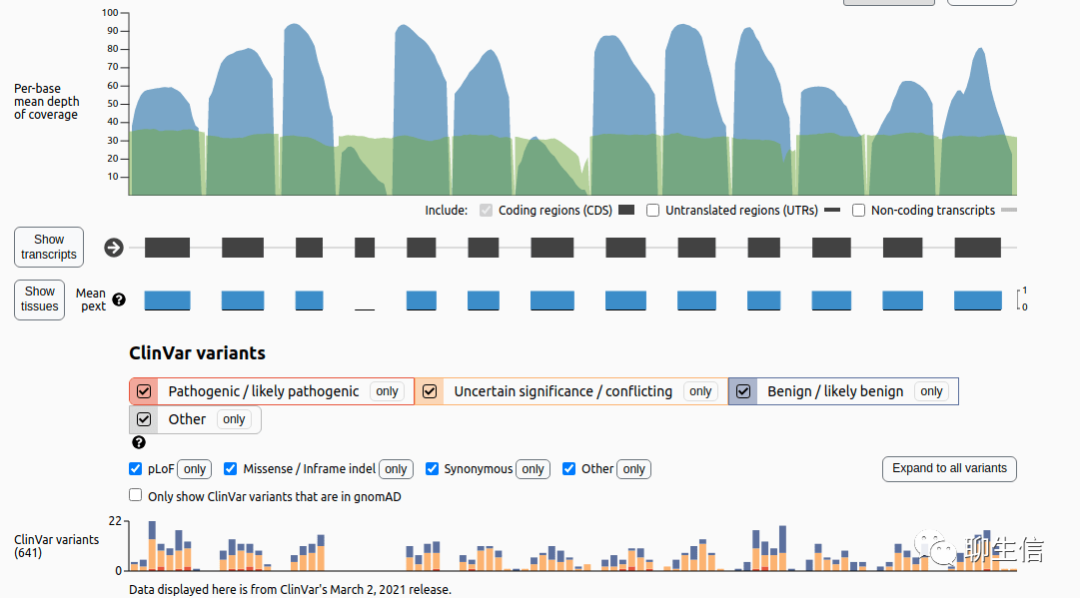
Genoverse
https://github.com/wtsi-web/Genoverse
Tags: General, Linear
Platform: Web
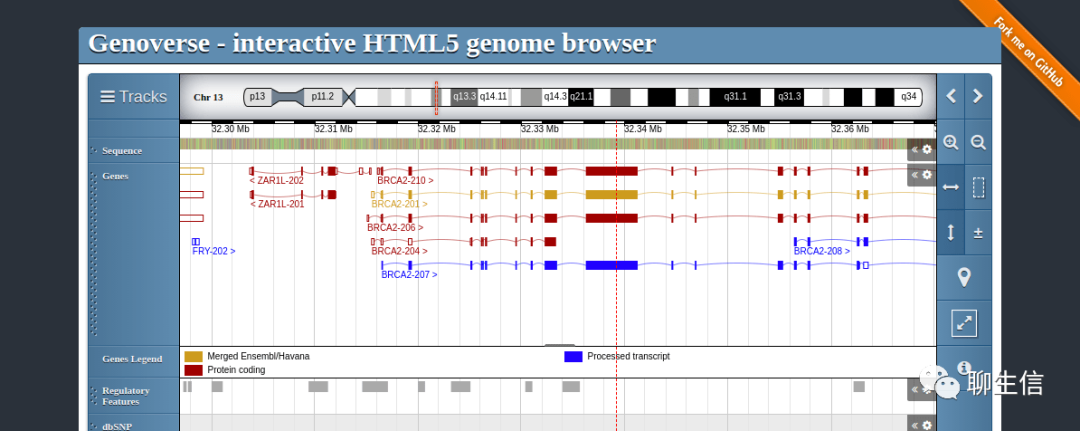
GenomeSpy
https://genomespy.app/
Language: JS, WebGL
Tags: CNV, GWAS, Linear, MSA
Platform: Web
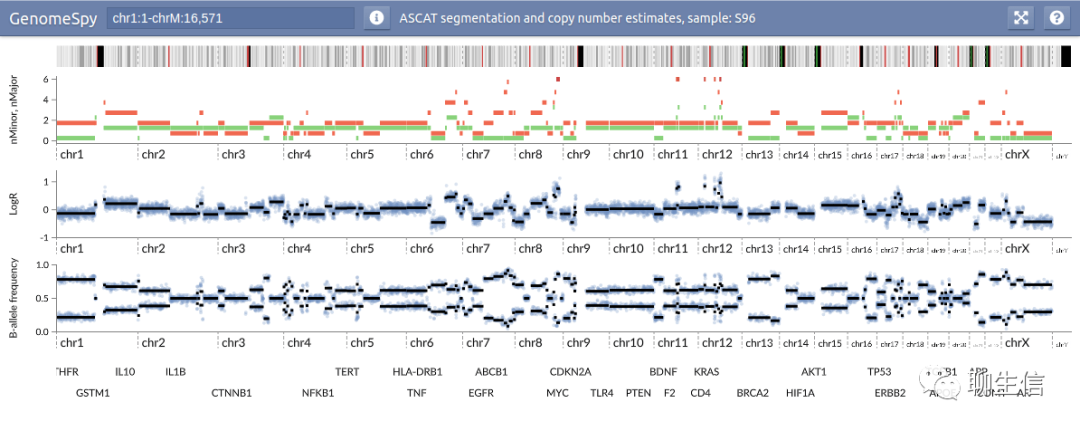
Ribbon
http://genomeribbon.com/
Publication: https://academic.oup.com/bioinformatics/advance-article-abstract/doi/10.1093/bioinformatics/btaa680/5885081?redirectedFrom=fulltext
Tags: SV, Alignment viewer
Github: https://github.com/MariaNattestad/ribbon
Platform: Web
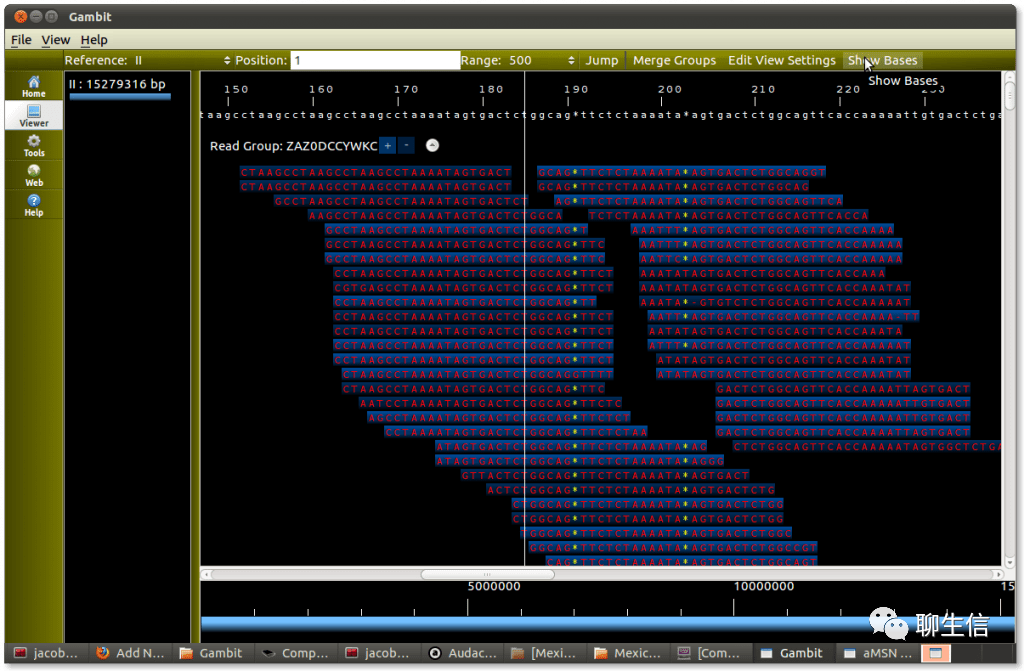
Gambit
http://labsergen.langebio.cinvestav.mx/bioinformatics/jacob/?p=473
Tags: Alignment viewer, Linear
Platform: Desktop
EaSeq
https://easeq.net/screenshots/
Publication: https://www.nature.com/articles/nsmb.3180
Tags: Special-purpose, Epigenomics
Platform: Desktop
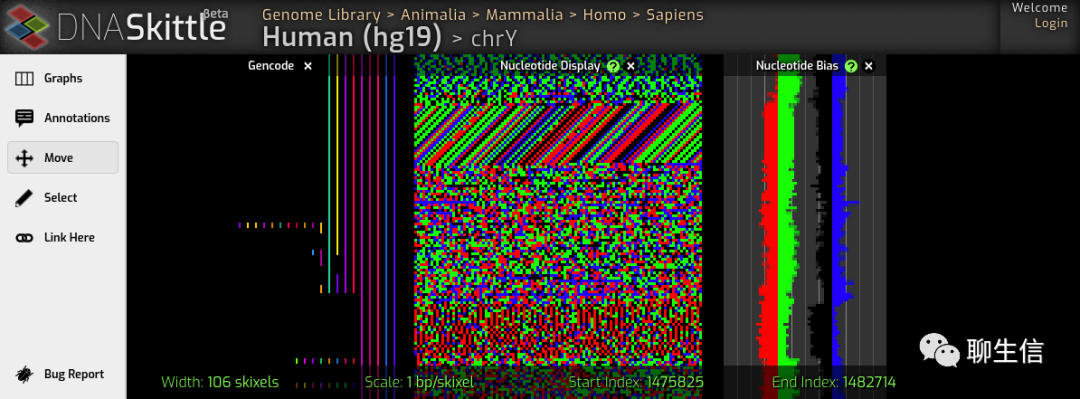
DNASkittle
https://www.dnaskittle.com/
Tags: Special-purpose
Platform: Web
D3GB
http://d3gb.usal.es/
Language: JS, D3
Tags: Special-purpose
Platform: Web
cnvCurator
http://www.acsu.buffalo.edu/~lm69/cnvCurator
Publication: http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0766-y
Language: Java
Tags: CNV, Annotation, Deadlink?
Platform: Desktop
Chromatic
https://chromatic.nci.nih.gov/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5987889/
Language: WASM
Tags: Alignment viewer
Platform: Web
Chopchop
http://chopchop.cbu.uib.no/results/1611256682447.8318/details/9
Tags: Synthetic biology
Apollo aka WebApollo
https://genomearchitect.readthedocs.io/en/latest/
Language: Java, Groovy, JS
Tags: Gene structure, Annotation, JBrowse integration
Platform: Web
Apollo
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC151184/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC151184/
Language: Java
Tags: Special-purpose, Historical, Deadlink?, Gene structure, Annotation, Comparative
Platform: Desktop
ApE (a plasmid editor)
https://jorgensen.biology.utah.edu/wayned/ape/
Tags: Synthetic biology
Platform: Desktop
seqviz
https://github.com/Lattice-Automation/seqviz#viewer
Tags: Synthetic biology
Platform: Web
OGDRAW
https://chlorobox.mpimp-golm.mpg.de/OGDraw.html
Publication: https://www.biorxiv.org/content/10.1101/545509v1.full.pdf
Tags: Circular, Static
Platform: Web
mummer2circos
https://github.com/metagenlab/mummer2circos
Language: Python
Tags: Static
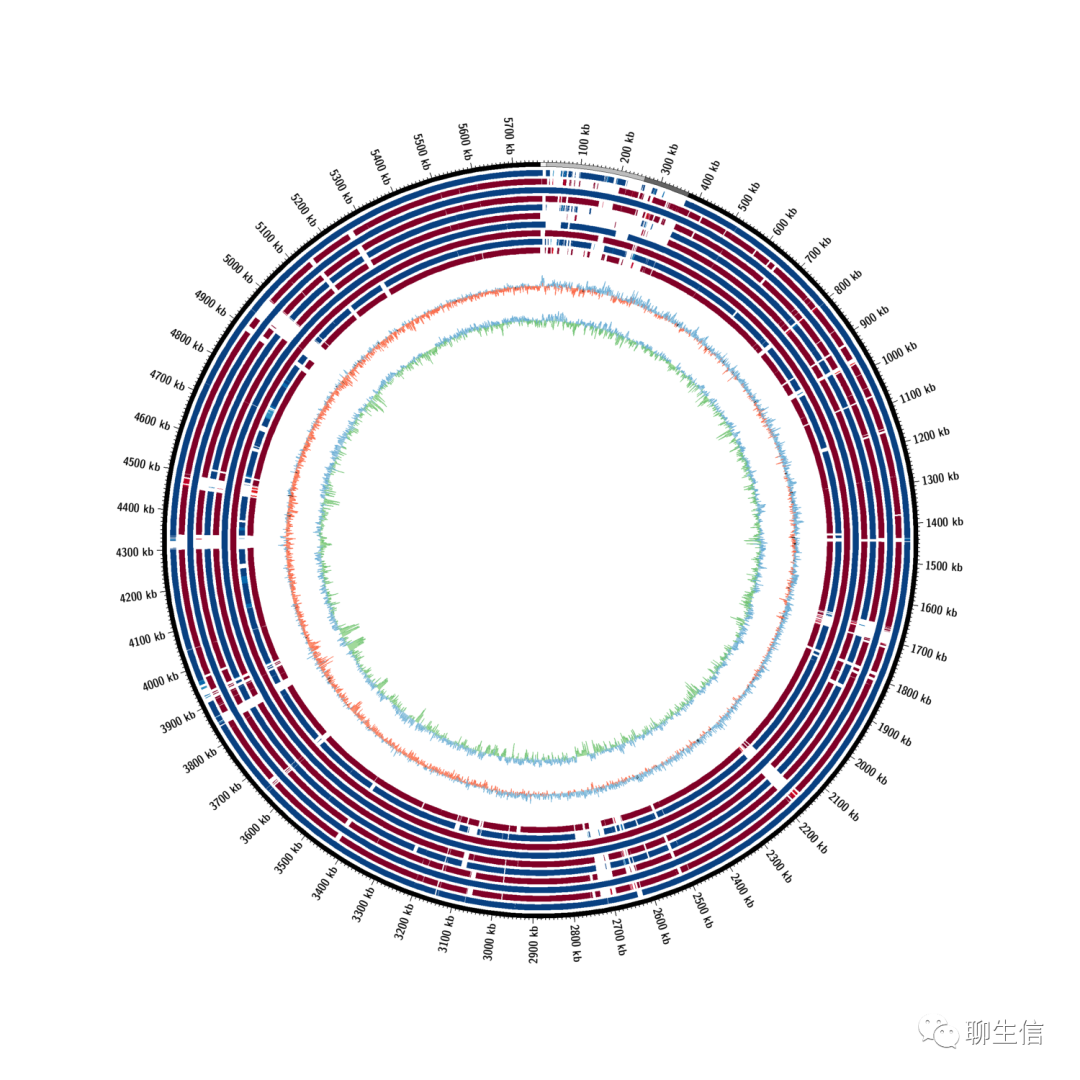
JupiterPlot
https://github.com/JustinChu/JupiterPlot
Language: Perl
Tags: Circular, Static, Comparative
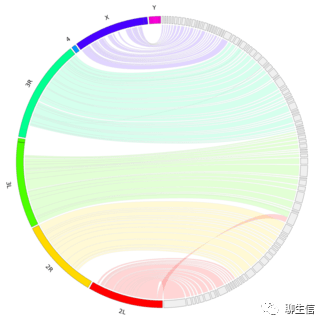
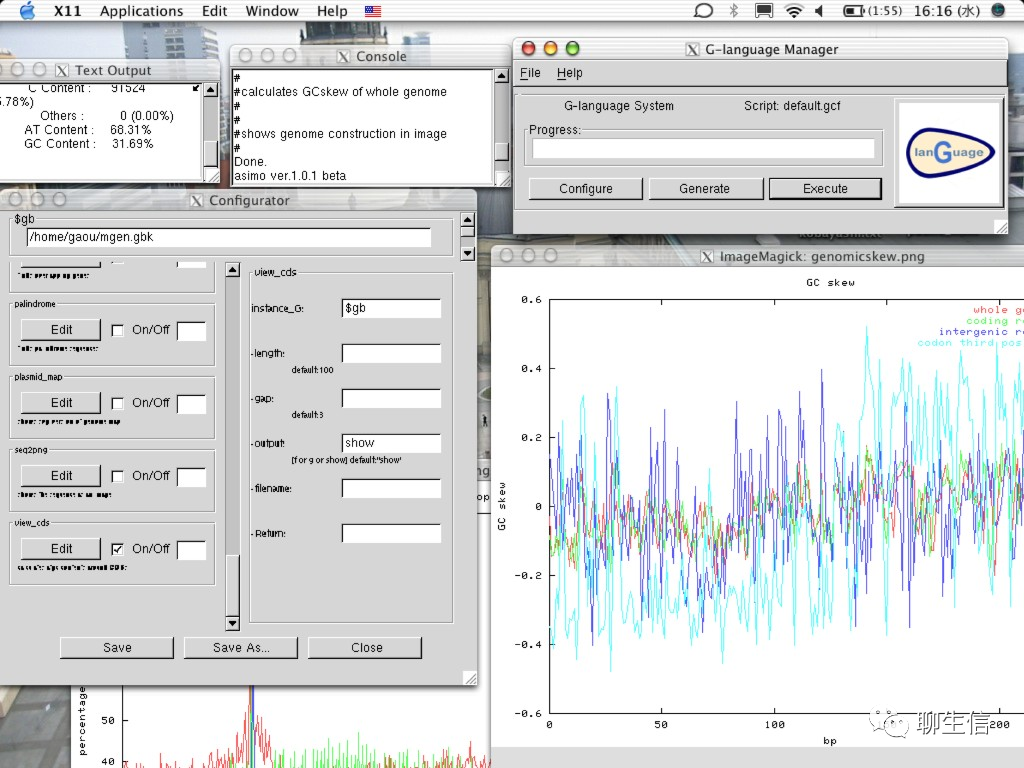
GenomeProjector
https://github.com/gaou/g-language/wiki
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2636772/
Language: Perl
Tags: Circular, Linear
Platform: Desktop
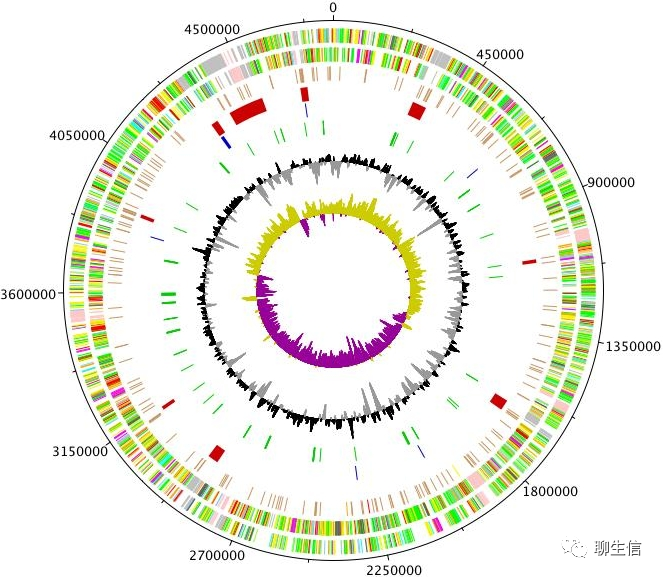
DNAPlotter
https://www.sanger.ac.uk/science/tools/dnaplotter
Language: Java
Tags: Circular
Platform: Desktop
CircosJS
https://github.com/nicgirault/circosJS
Language: JS
Tags: Circular
Platform: Web
Circleator
http://jonathancrabtree.github.io/Circleator/index.html
Language: Perl
Tags: Circular, Static
Circa
https://omgenomics.com/circa
Tags: Commercial, Circular
Platform: Desktop
BioCircos.js
http://bioinfo.ibp.ac.cn/biocircos/
Language: JS
Tags: Circular
Platform: Web
VizAln (from HipSTR)
https://github.com/tfwillems/HipSTR
Tags: Text based
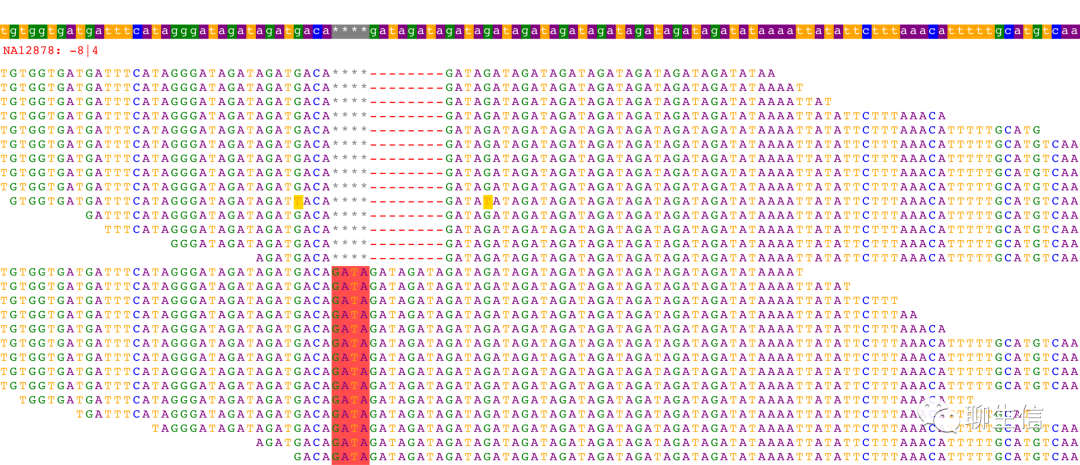
SvABA
https://github.com/walaj/svaba#alignmentstxtgz
Tags: Text based
Platform: CLI

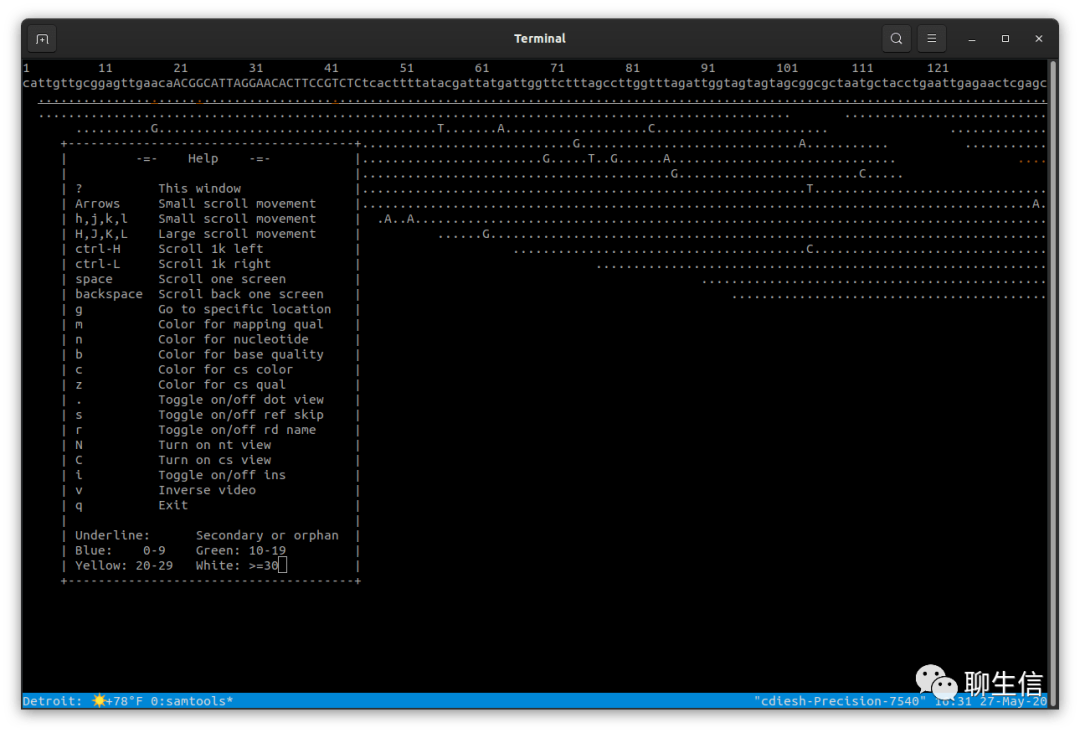
Samtools tview
http://www.htslib.org/
Language: C
Tags: Text based, Alignment viewer
Platform: CLI
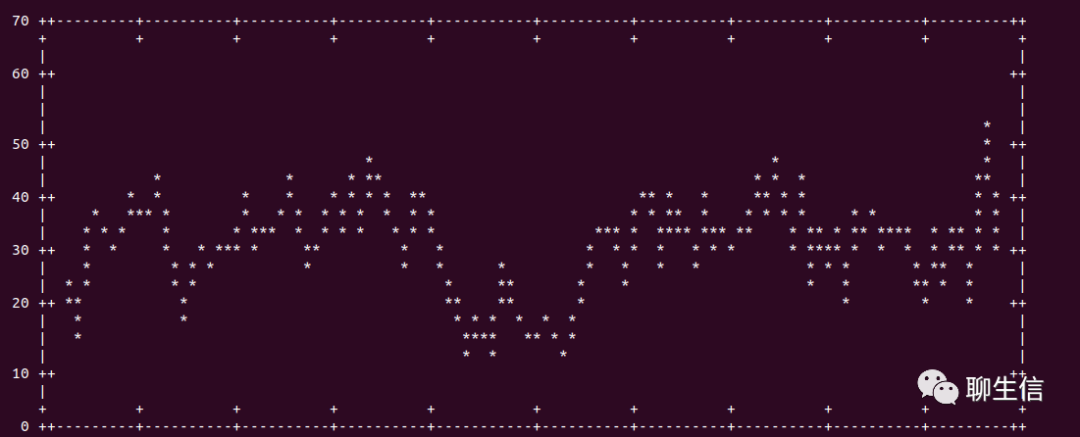
Samtools depth visualization
https://twitter.com/yokofakun/status/1178686978541441025
Tags: Text based, Coverage
Platform: CLI, GNUplot
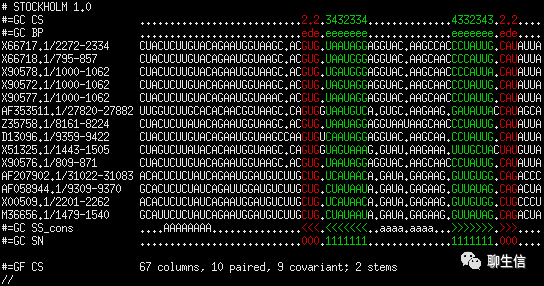
Rna Alignment Viewers
http://biowiki.org/wiki/index.php?title=Rna_Alignment_Viewers&redirect=no
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7109877/
Language: Perl
Tags: Text based
Platform: CLI
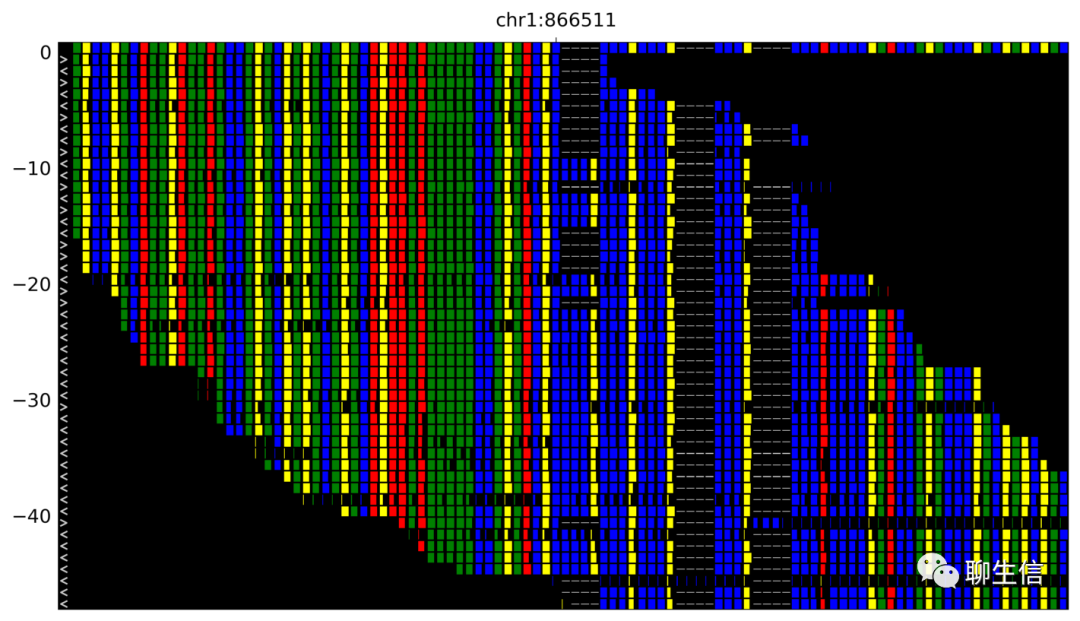
pyBamView
https://mgymrek.github.io/pybamview/
Language: Python
Tags: Alignment viewer, Static
Note: See supplementary info for more figures, supports padded SAM/BAM which is fairly rare
Platform: CLI
plotReads
http://campuspress.yale.edu/knightlab/ruddle/plotreads/
Tags: Text based
Platform: CLI, Static
Hapviz
https://github.com/ekg/hapviz
Language: C
Tags: Text based
Platform: CLI

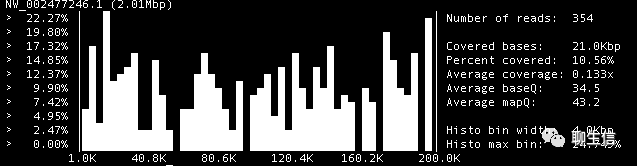
Bamcov
https://github.com/fbreitwieser/bamcov
Language: C
Tags: Text based, Coverage
Platform: CLI
ASCIIGenome
https://github.com/dariober/ASCIIGenome
Language: Java
Tags: Text based, Alignment viewer
Platform: CLI
Alv
https://github.com/arvestad/alv
Language: Python
Tags: Text based, MSA
Platform: CLI
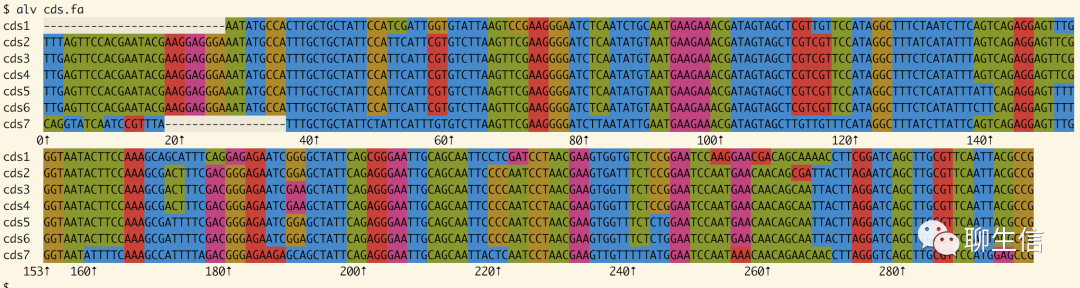
Alan
https://github.com/mpdunne/alan
Language: Bash
Tags: Text based, MSA
Platform: CLI
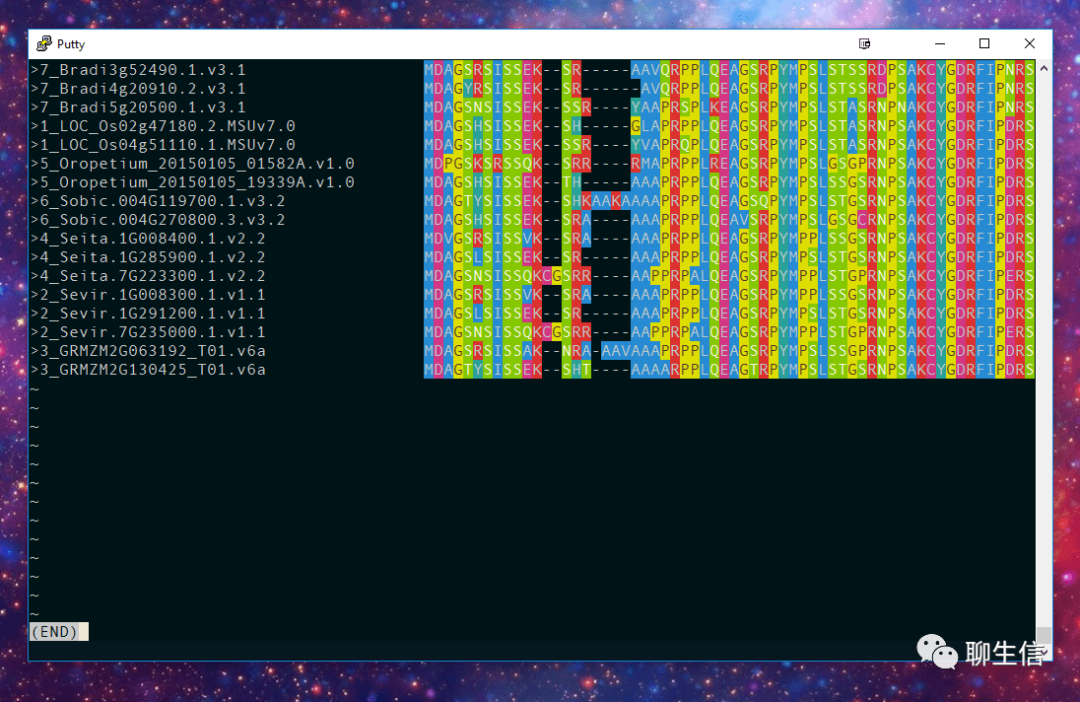
sequence tube map
https://github.com/vgteam/sequenceTubeMap
Language: JS
Tags: Graph
Platform: Web
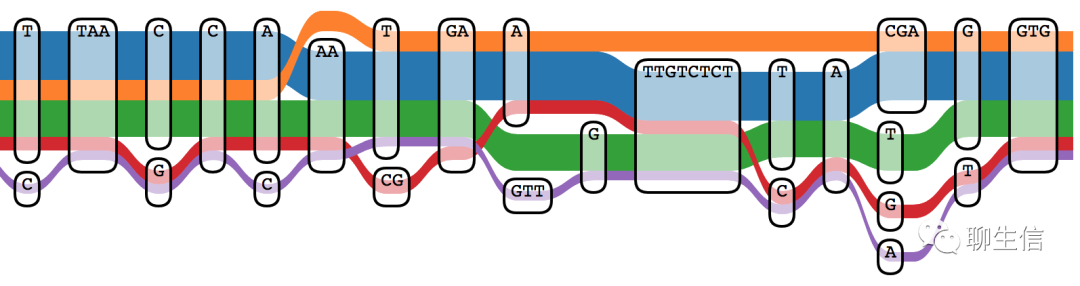
odgi draw + odgi viz
https://odgi.readthedocs.io/en/latest/index.html
Tags: Graph
Note: Example from https://github.com/pangenome/pggb
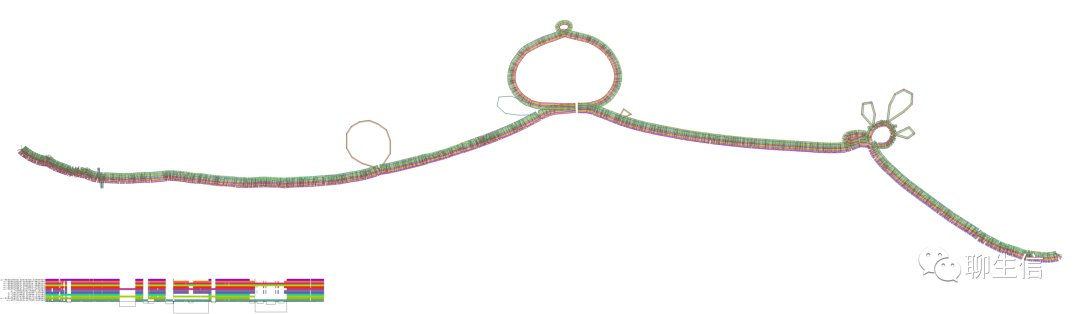
MoMi-G
https://github.com/MoMI-G/MoMI-G/
Language: JS
Tags: Graph, Multi, Alignment viewer, Circular, SV
Platform: Web
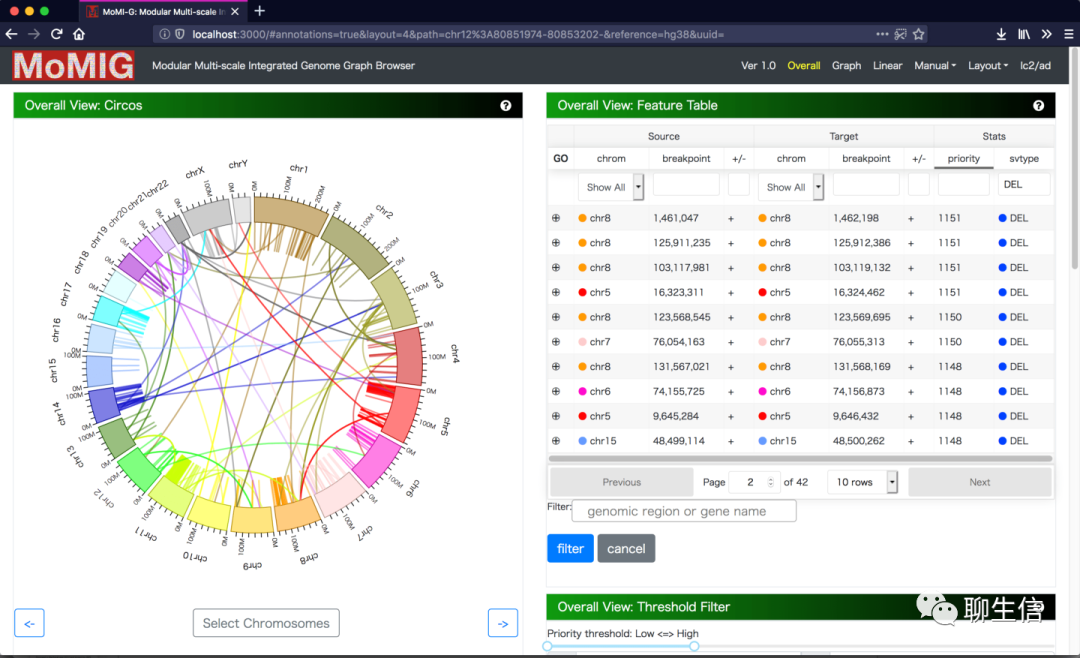
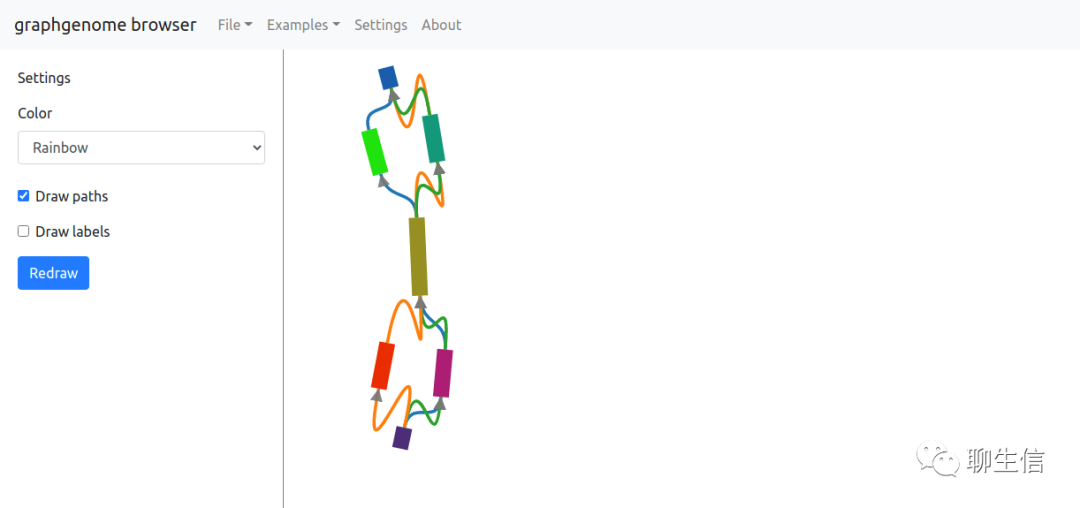
graphgenomeviewer
https://cmdcolin.github.io/graphgenomeviewer/
Language: JS
Tags: Graph
Platform: Web
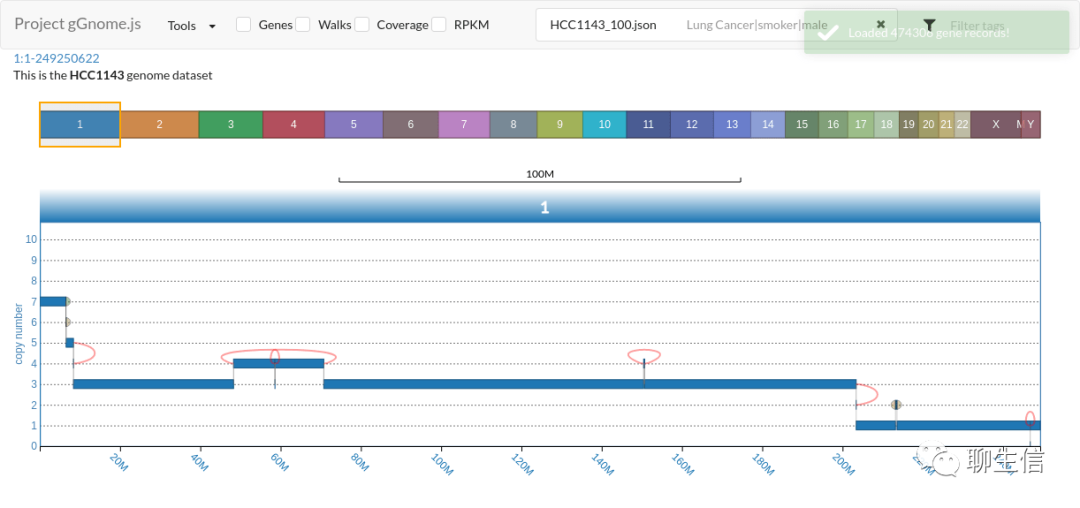
gGnomes.js
https://github.com/mskilab/gGnome.js
Language: JS
Tags: SV, CNV, Breakends, Graph
Platform: Web
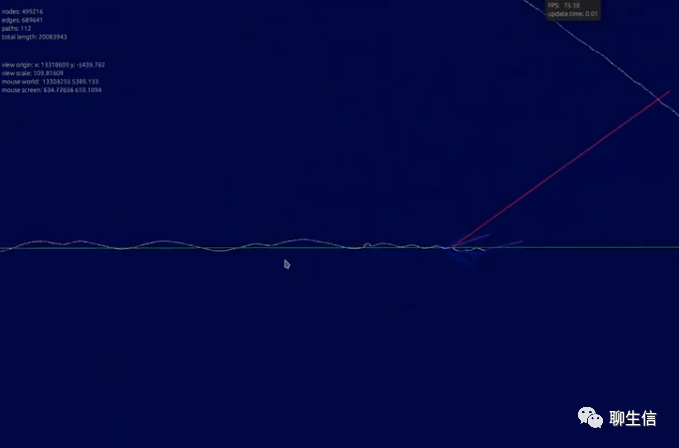
gfaestus
https://github.com/chfi/gfaestus
Language: Rust
Tags: Graph
Note: See demo video displaying GFF3 annotations on graph https://www.youtube.com/watch?v=A-HnKXIrJl4
Bandage
https://github.com/rrwick/Bandage/
Publication: https://academic.oup.com/bioinformatics/article/31/20/3350/196114
Language: C++
Tags: Graph
Platform: Desktop
XMatchView
https://www.bcgsc.ca/resources/software/xmatchview
Tags: Comparative
Platform: Desktop
VISTA browser
http://pipeline.lbl.gov/cgi-bin/gateway2
Tags: Comparative
Platform: Web, Silo
Tripal MapViewer
https://academic.oup.com/database/article/doi/10.1093/database/baz100/5612723
Tags: Comparative, Ideogram
SynVisio
https://synvisio.github.io/
Language: JS
Tags: Comparative, Linear, Dotplot

SynTView
http://hub18.hosting.pasteur.fr/SynTView/documentation/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3849071/
Tags: Comparative, Dotplot, Linear
Platform: Desktop, Web, Flash
Synteny browser
https://github.com/TheJacksonLaboratory/syntenybrowser
Tags: Comparative
Platform: Web
Synima
https://github.com/rhysf/Synima
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5697234/
Language: Perl
Tags: Comparative
Platform: CLI
SyMap
http://www.agcol.arizona.edu/software/symap/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3105427/
Tags: Comparative, Dotplot, Linear
Platform: Desktop
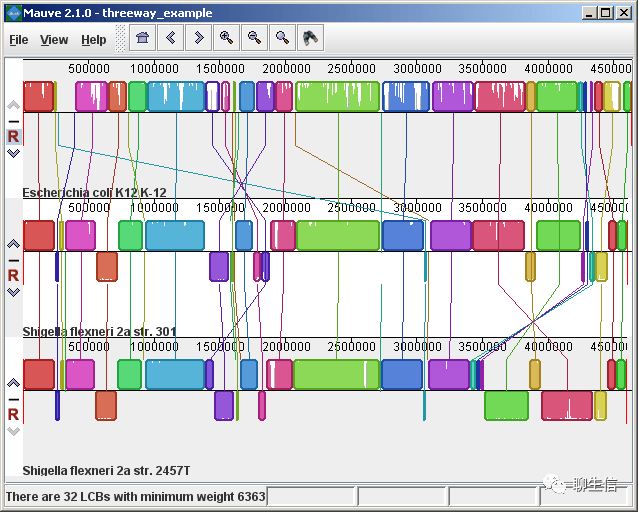
progressiveMauve viewer
http://darlinglab.org/mauve/user-guide/viewer.html
Tags: Comparative
Platform: Desktop
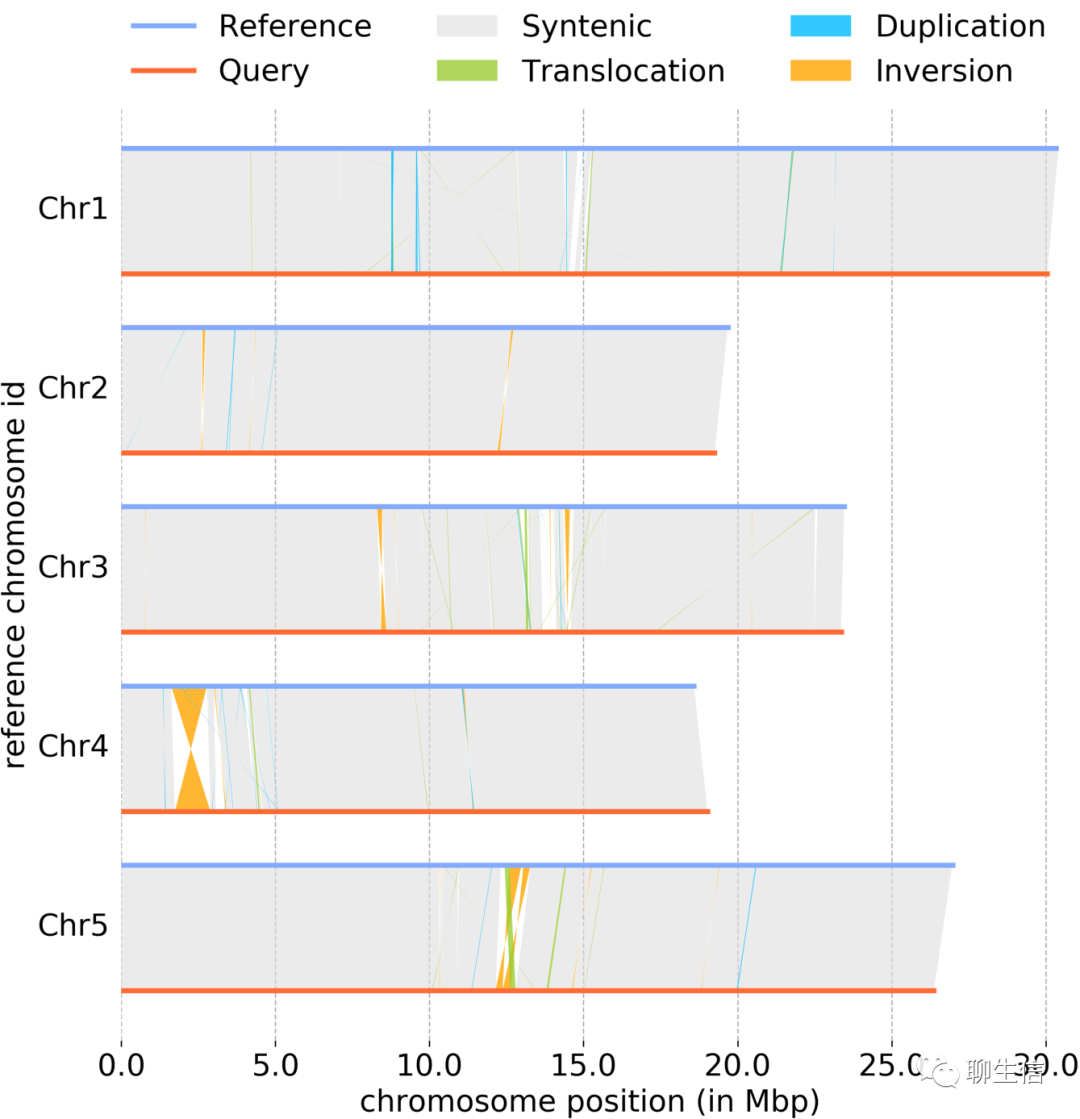
syri/plotsr
https://schneebergerlab.github.io/syri/plotsr.html
Language: Python
Tags: Comparative, Linear
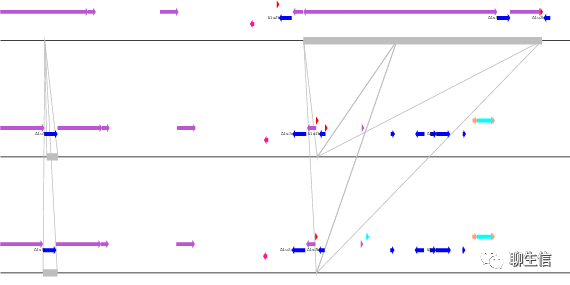
Parasight
https://baileylab.brown.edu/parasight/galframe.html
Language: Perl
Tags: Comparative
Note: Examples at https://ratparalogy.gs.washington.edu/
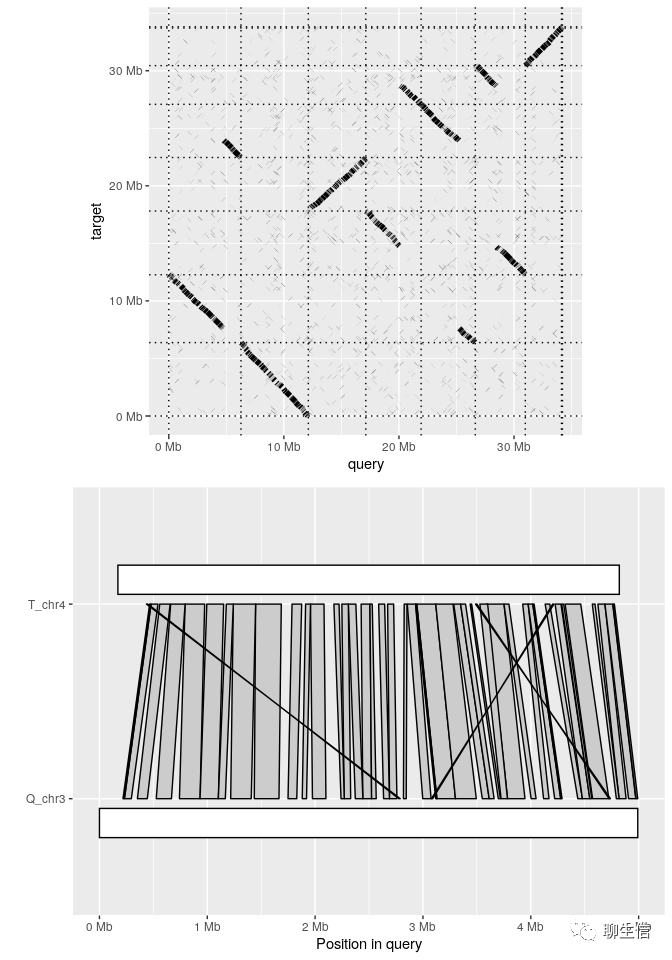
pafr
https://github.com/dwinter/pafr
Language: R
Tags: Comparative, Dotplot, Linear
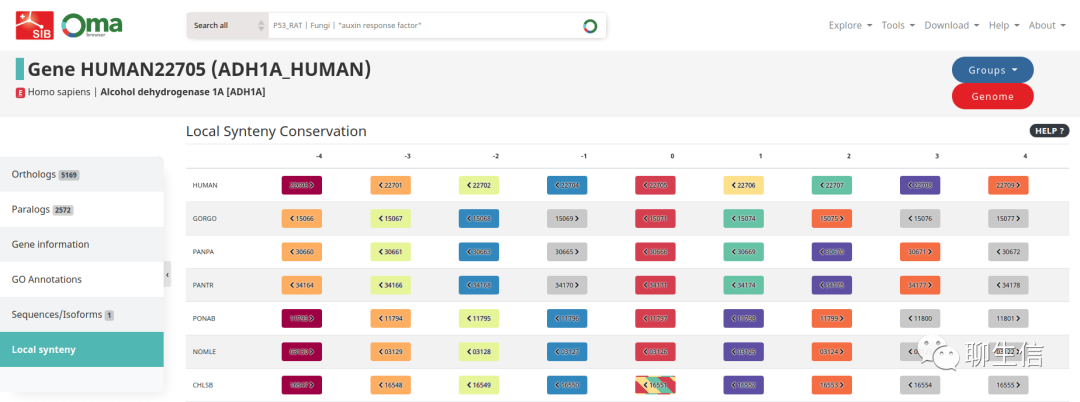
OMA local synteny browser
https://omabrowser.org/oma/synteny/ADH1A_HUMAN/
Tags: Comparative, Gene order
Platform: Web, Silo
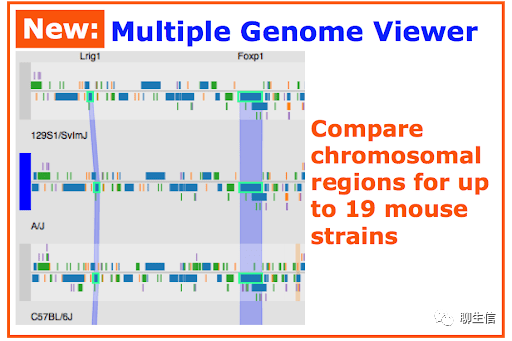
Multiple genome viewer
http://www.informatics.jax.org/mgv/
Tags: Comparative
Platform: Web, Silo
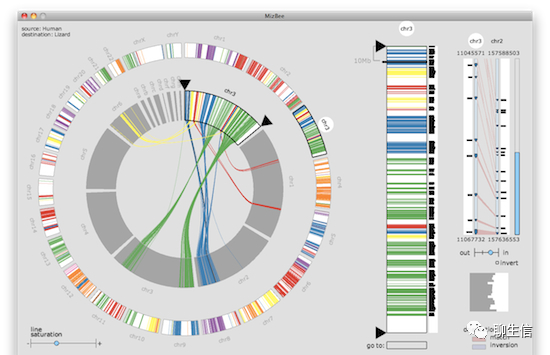
MizBee
http://www.cs.utah.edu/~miriah/mizbee/Overview.html
Publication: https://ieeexplore.ieee.org/document/5290692
Language: Processing
Tags: Comparative
Platform: Desktop
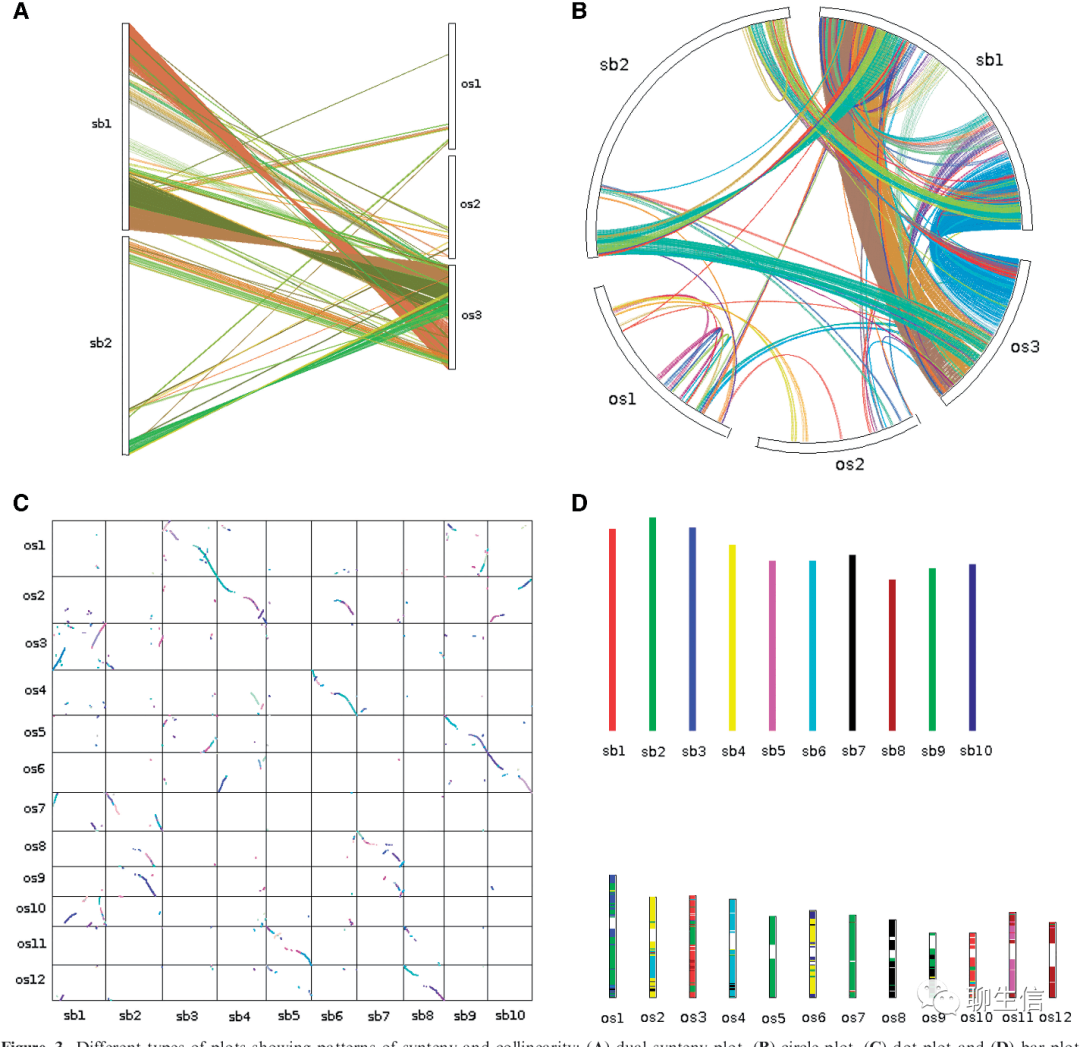
MCScanX
https://github.com/wyp1125/MCScanX
Language: Perl
Tags: Comparative, Dotplot, Circular, Linear, Ideogram
Platform: CLI
MCScan (python version)
https://github.com/tanghaibao/jcvi/wiki/MCscan-(Python-version)
Language: Python
Tags: Comparative, Dotplot, Linear
Platform: CLI
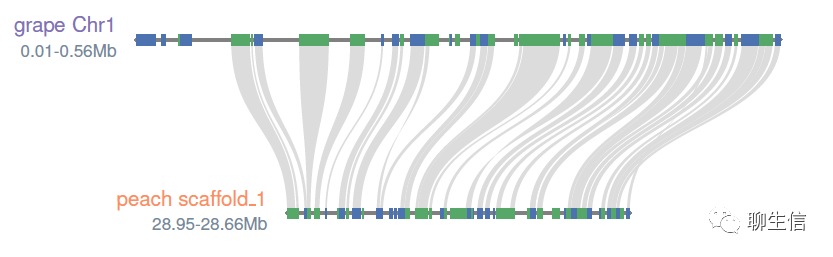
mauve-viewer
https://github.com/nconrad/mauve-viewer
Language: JS
Tags: Comparative
Platform: Web
GIVE
https://zhong-lab-ucsd.github.io/GIVE_homepage/
Language: JS
Tags: Comparative, Linear, Quantitative
Platform: Web
gggenomes
https://github.com/thackl/gggenomes
Language: R
Tags: Comparative
Platform: Package
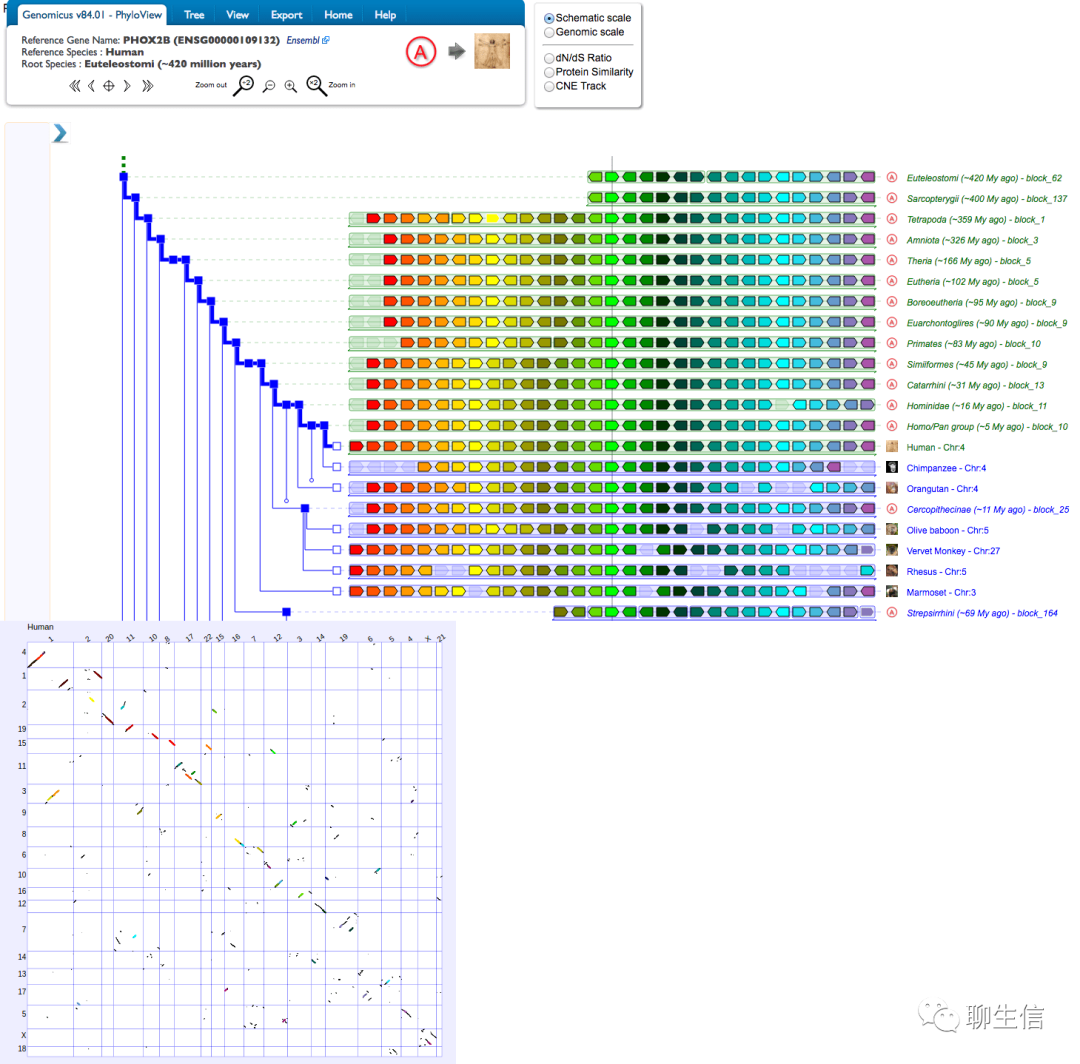
Genomicus
https://www.genomicus.biologie.ens.fr/genomicus/
Tags: Comparative, Ideogram, Dotplot
Platform: Web, Silo
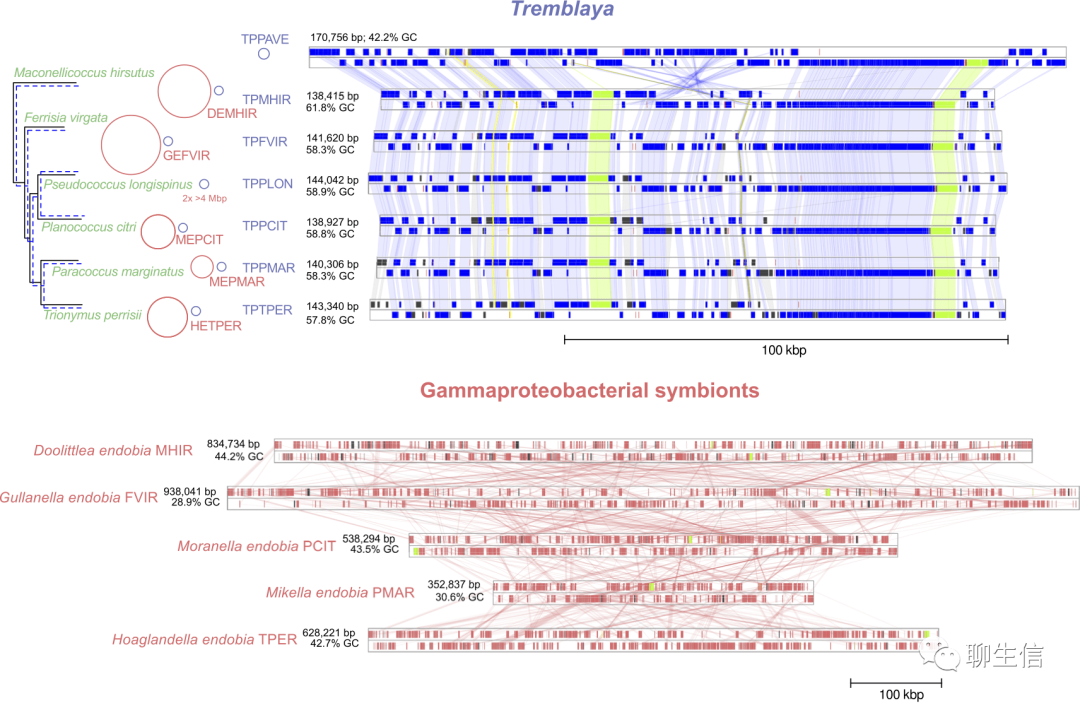
Genome-plots-processing
https://github.com/filip-husnik/genome-plots-processing
Language: Processing
Tags: Comparative
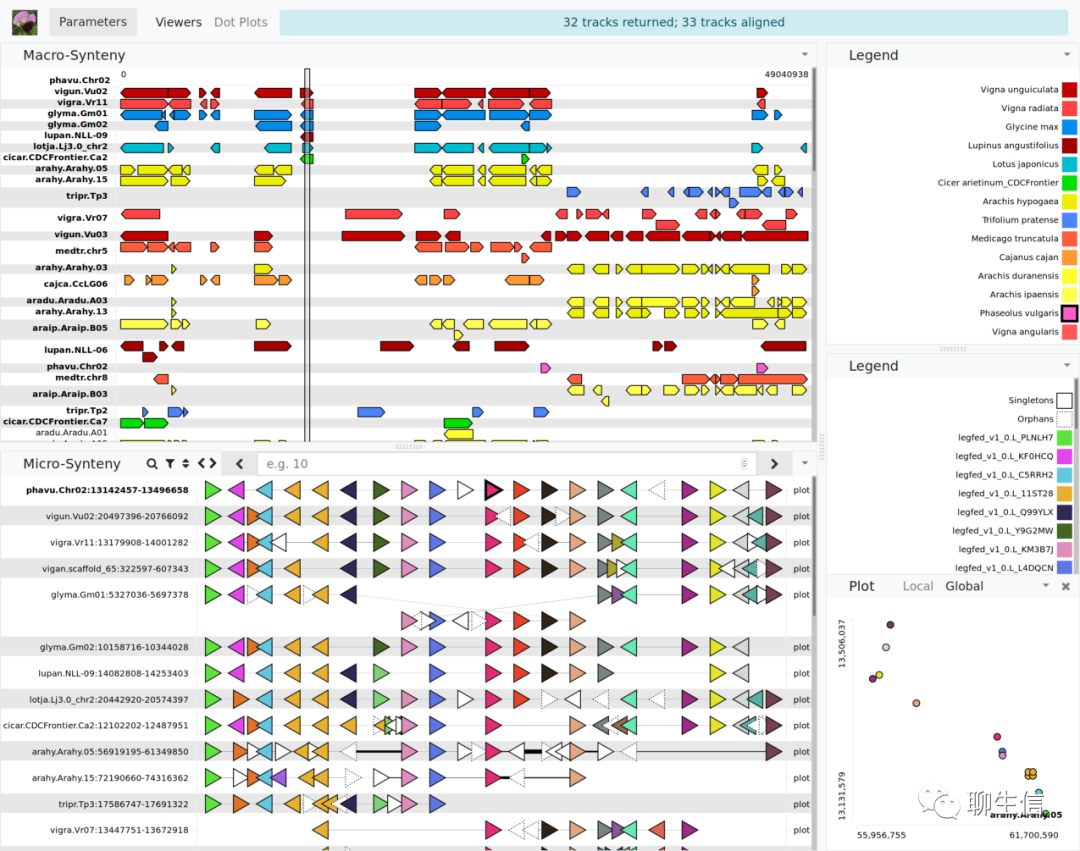
Genome Context Viewer
https://www.legumefederation.org/gcv/phytozome_10_2/
Tags: Comparative, Gene order
Platform: Web, Silo
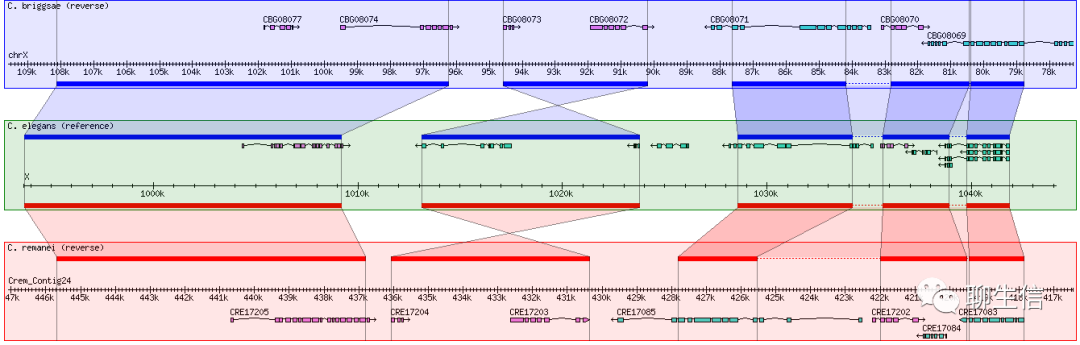
GBrowse_syn
http://gmod.org/wiki/GBrowse_syn
Language: Perl
Tags: Comparative
Platform: Web
Edinburgh-Genome-Foundry/DnaFeaturesViewer
https://github.com/Edinburgh-Genome-Foundry/DnaFeaturesViewer
Publication: https://academic.oup.com/bioinformatics/article-abstract/36/15/4350/5868559?redirectedFrom=PDF
Language: Python
Tags: Microbiology, Synthetic biology
Platform: Desktop
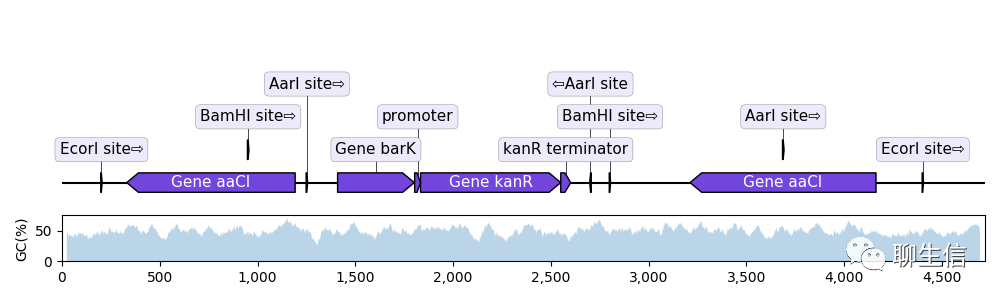
EasyFig
https://mjsull.github.io/Easyfig/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3065679/
Language: Python
Tags: Comparative
Platform: CLI
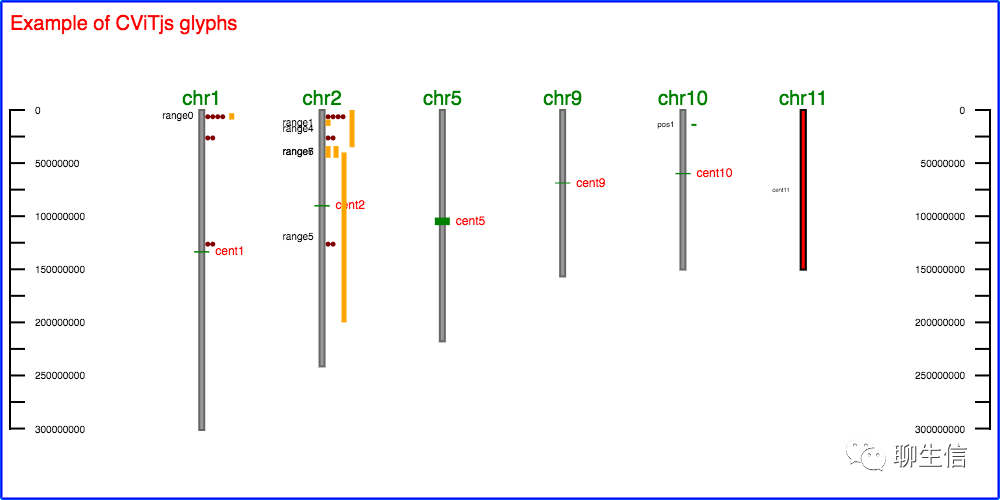
Cvit.js
https://github.com/LegumeFederation/cvitjs
Tags: Comparative
Platform: Web
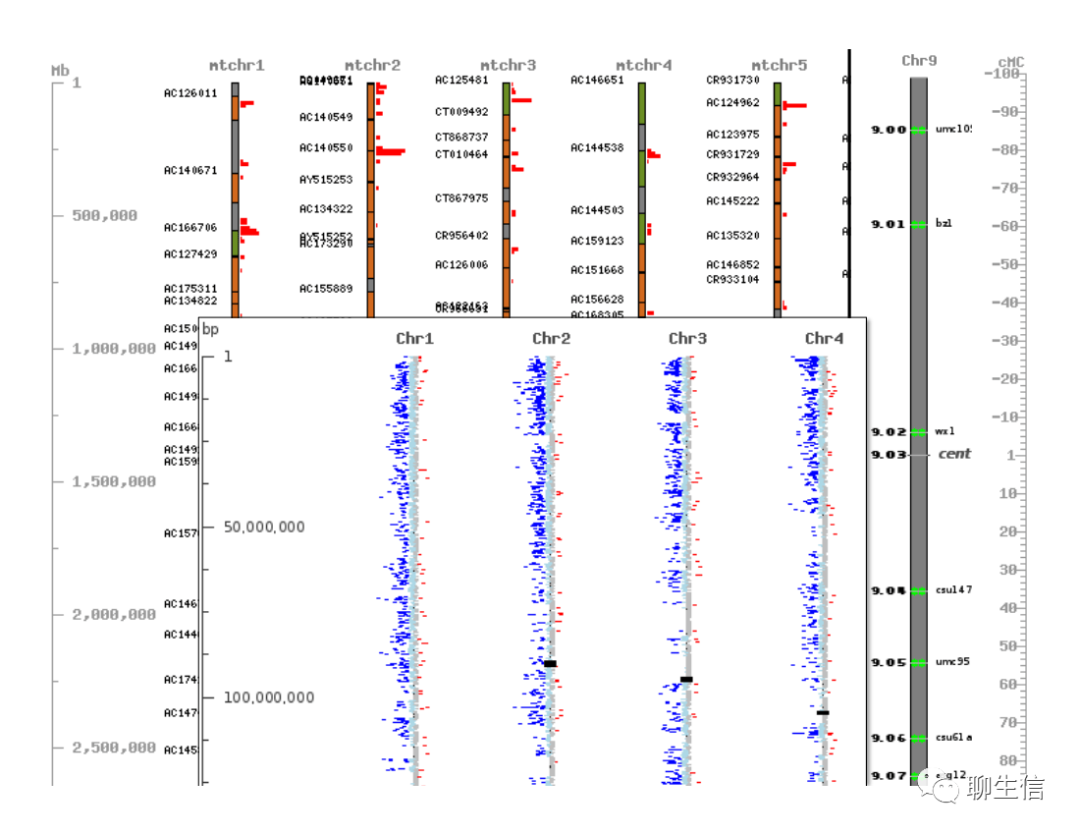
CVit
https://sourceforge.net/projects/cvit/
Language: Perl
Tags: Comparative
Platform: Web
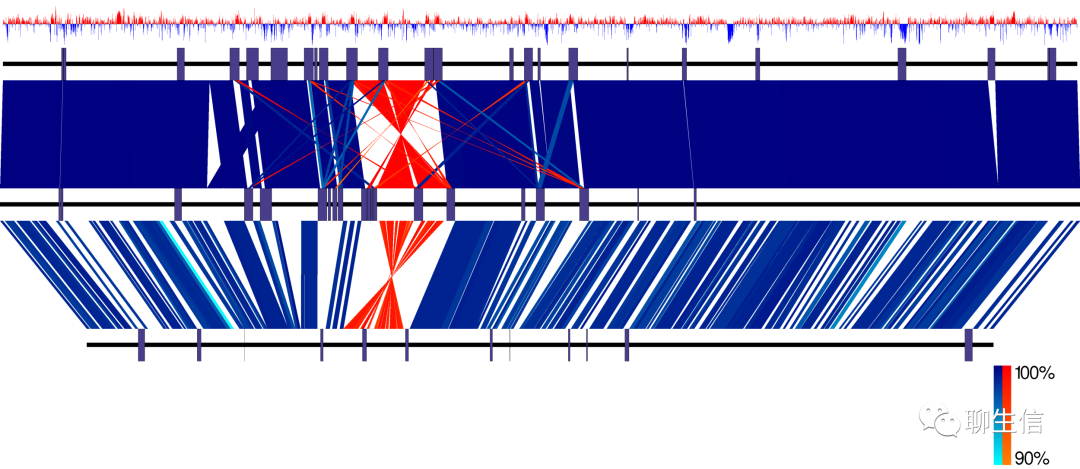
CrossBrowse
https://github.com/shenkers/CrossBrowse
Publication: https://www.biorxiv.org/content/10.1101/272880v1.full
Language: Java
Tags: Comparative
Platform: Desktop
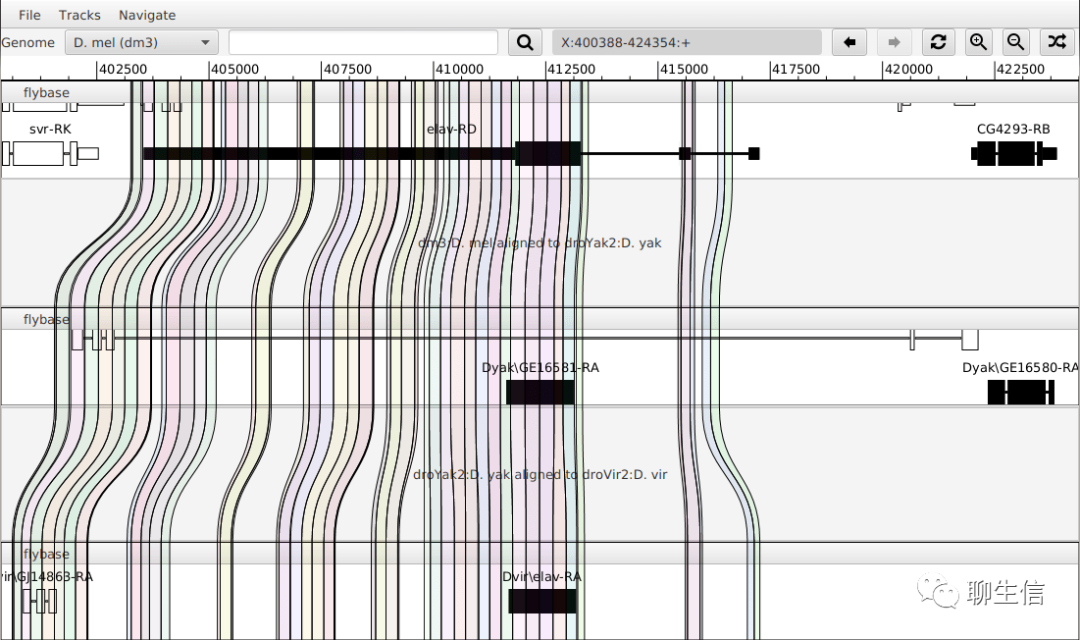
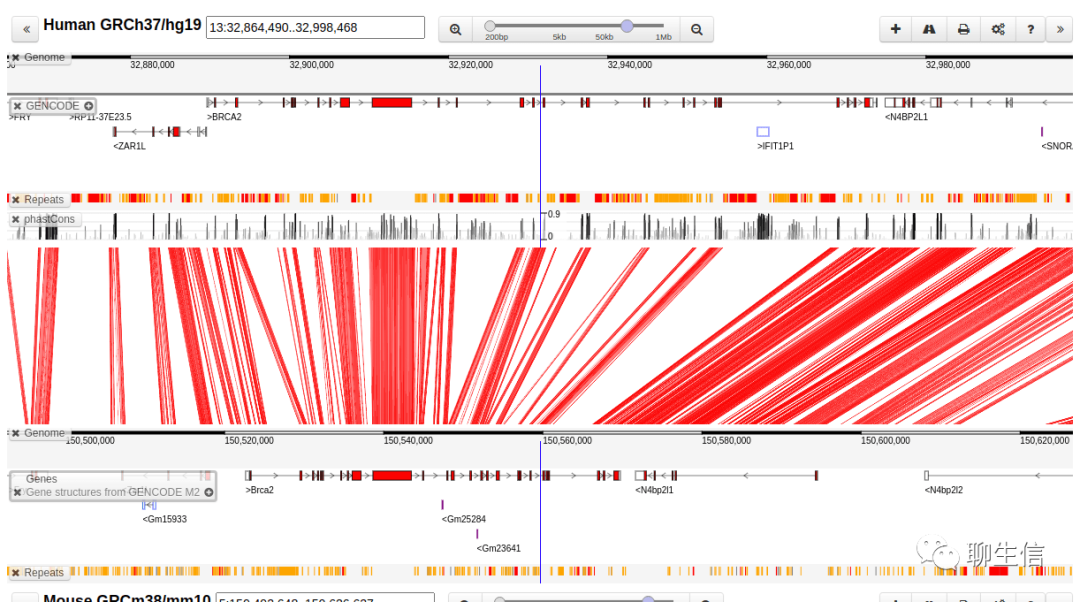
Comparative assembly hub / "snake track"
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4296145/
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4296145/
Tags: Comparative, UCSC integration
Platform: Web
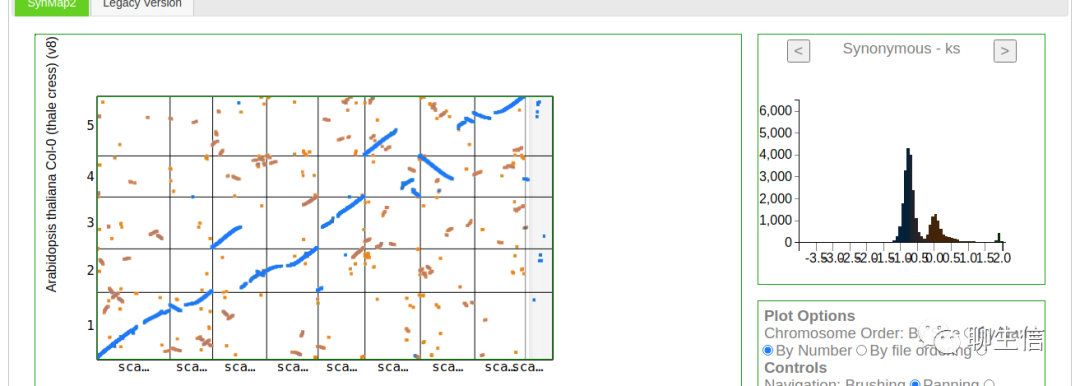
CoGe
https://genomevolution.org/coge/
Language: Perl
Tags: Comparative, Dotplot, Linear, JBrowse integration
Platform: Web, Silo
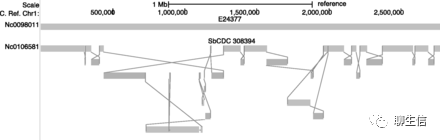
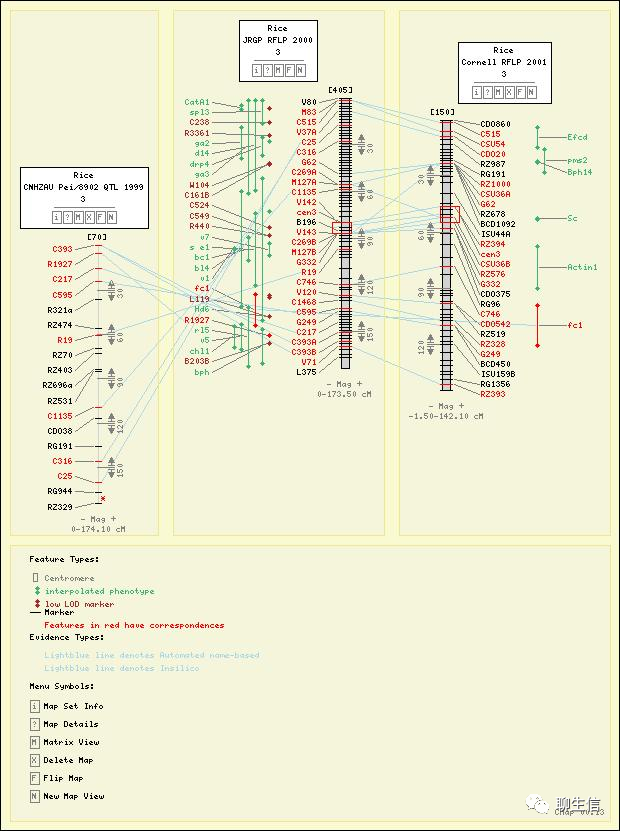
CMap
http://gmod.org/wiki/CMap
Language: Perl
Tags: Comparative
Platform: Web
Chromatiblock
https://github.com/mjsull/chromatiblock
Publication: https://www.biorxiv.org/content/10.1101/800920v2.full.pdf
Language: Python
Tags: Comparative, Pangenome
Platform: Web
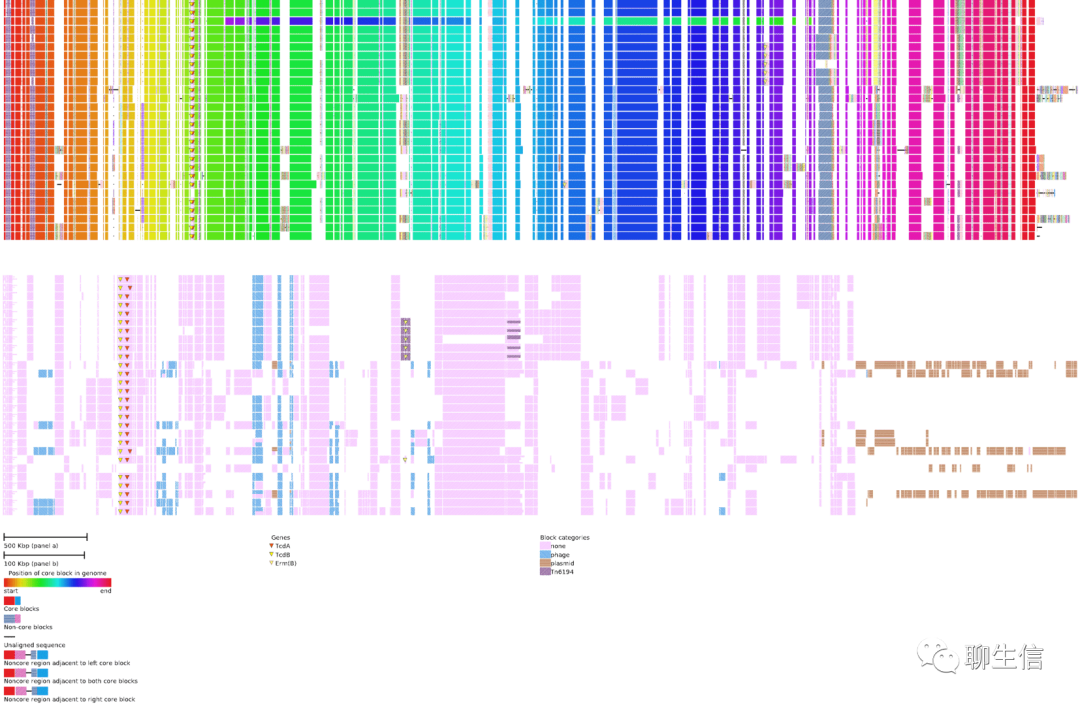
Biodalliance comparative demo
http://biodalliance.org/dev/test-comparative.html
Language: JS
Tags: Comparative
Platform: Web
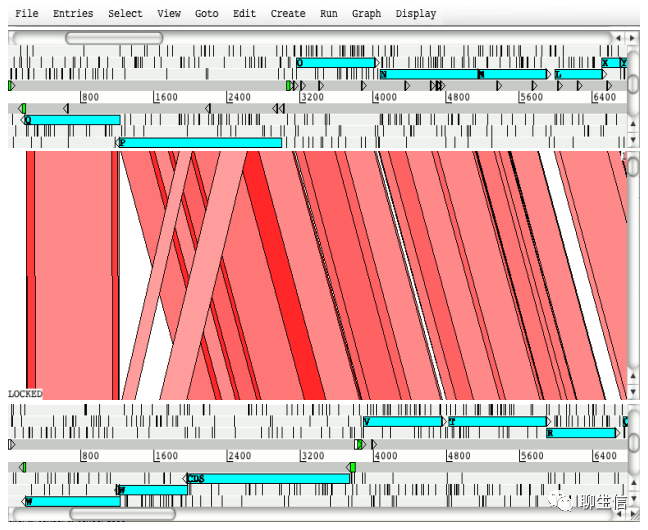
Artemis comparison tool (ACT)
https://www.sanger.ac.uk/science/tools/artemis-comparison-tool-act
Language: Java
Tags: Comparative
Platform: Desktop
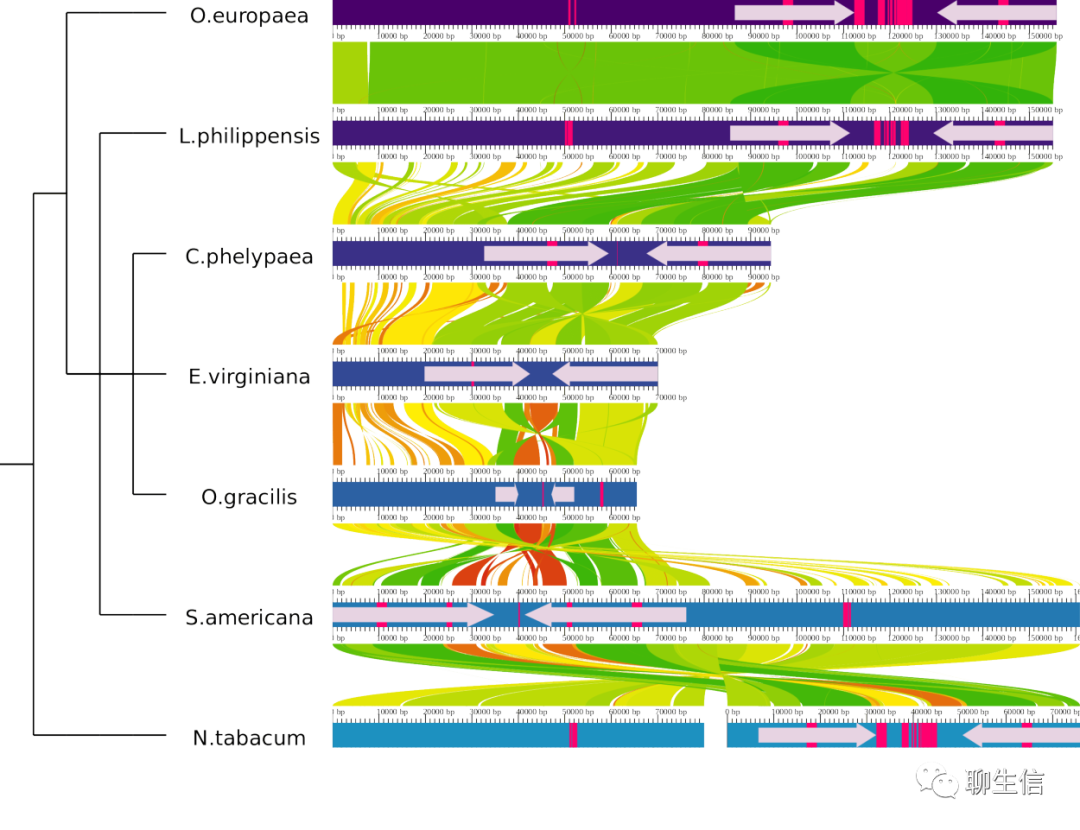
AliTV
https://alitvteam.github.io/AliTV/d3/AliTV.html
Tags: Comparative
Platform: Web
Aequatus
https://github.com/TGAC/Aequatus
Tags: Comparative, Deadlink?
Platform: Web, Silo

trackViewer
https://bioconductor.org/packages/release/bioc/vignettes/trackViewer/inst/doc/trackViewer.html
Language: R
Tags: Static, Lollipops, Quantitative
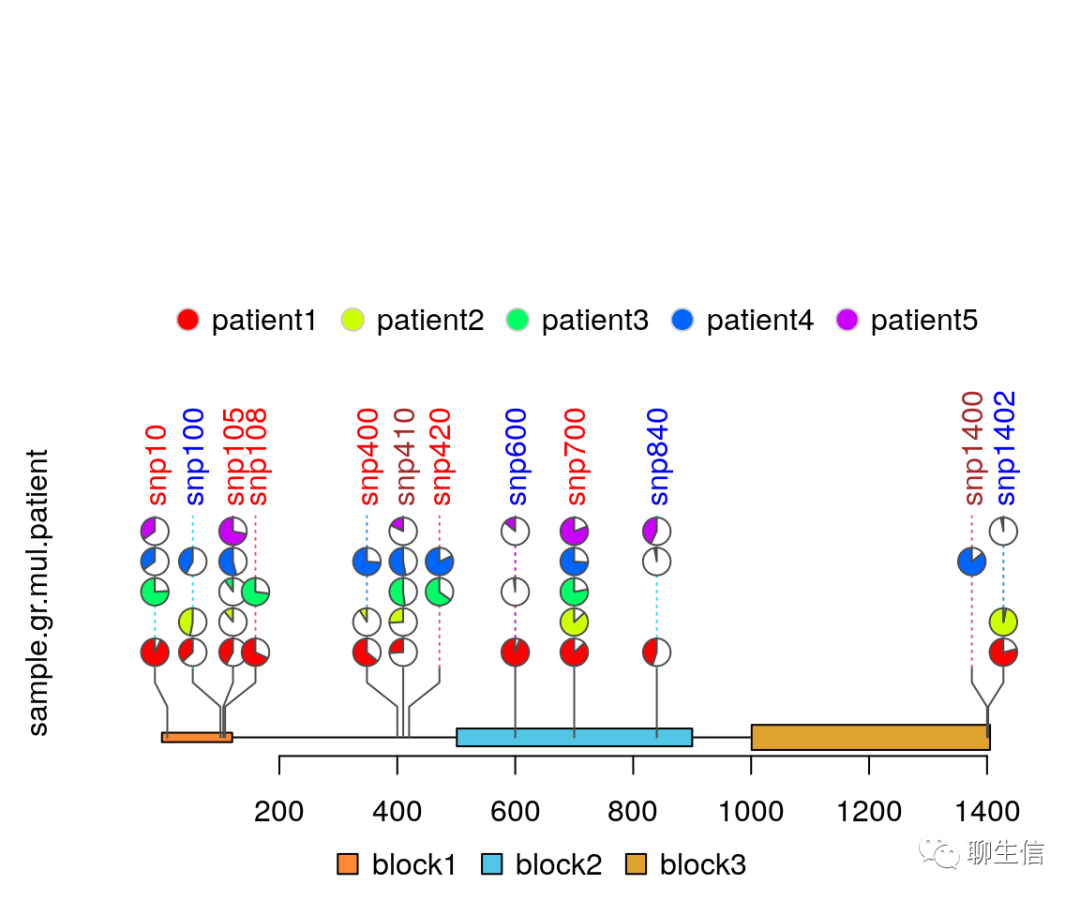
TnT
https://tnt.marlin.pub/articles/introduction
Language: R, JS
Tags: Gene structure
Platform: Web, Interactive
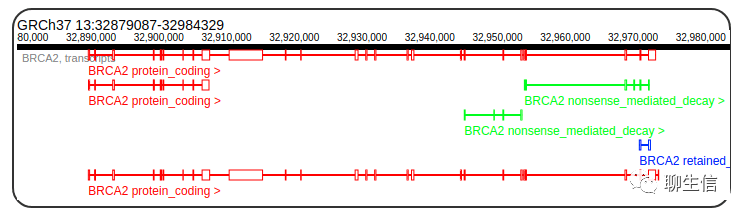
svviz2
https://github.com/nspies/svviz2
Publication: https://pubmed.ncbi.nlm.nih.gov/26286809/
Language: Python
Tags: Static, SV, Insertion
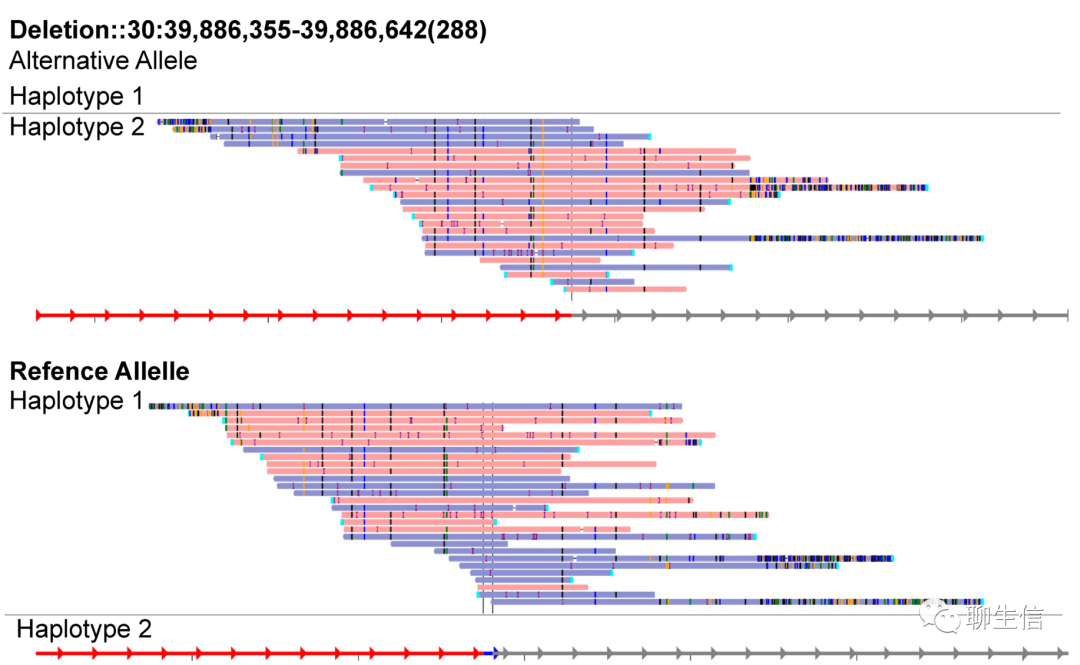
svviz
https://github.com/svviz/svviz
Publication: https://pubmed.ncbi.nlm.nih.gov/26286809/
Language: Python
Tags: Static, SV, Insertion
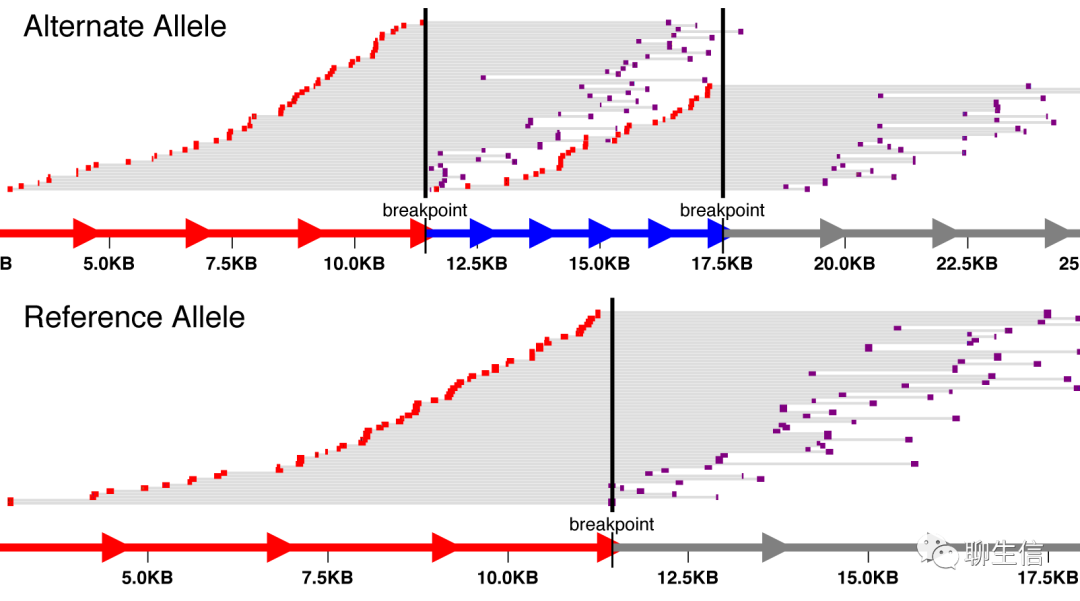
svv
https://github.com/ryanlayer/svv
Language: Python
Tags: Static, SV, Coverage
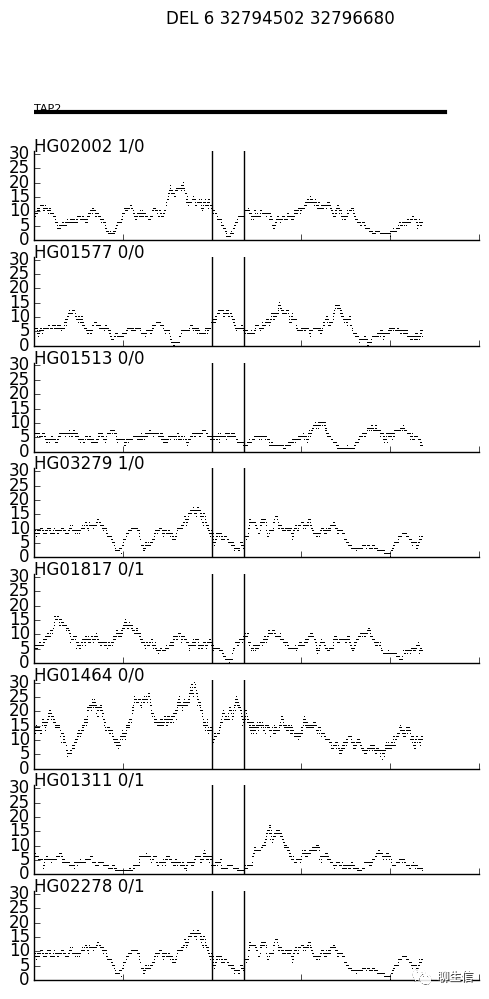
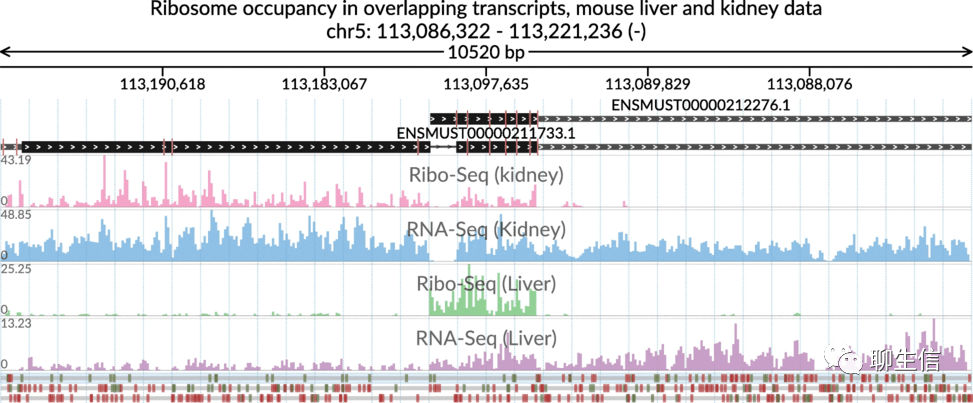
svist4get
https://link.springer.com/article/10.1186/s12859-019-2706-8
Publication: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-019-2706-8
Language: Python
Tags: Static
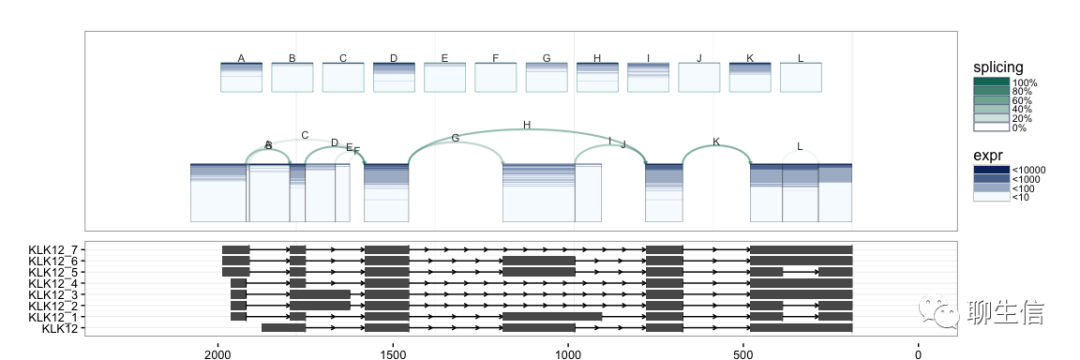
Sushi
https://www.bioconductor.org/packages/release/bioc/vignettes/Sushi/inst/doc/Sushi.pdf
Language: R
Tags: Static, Multi
Spliceclust
https://github.com/pkimes/spliceclust
Language: R
Tags: Static, Splicing
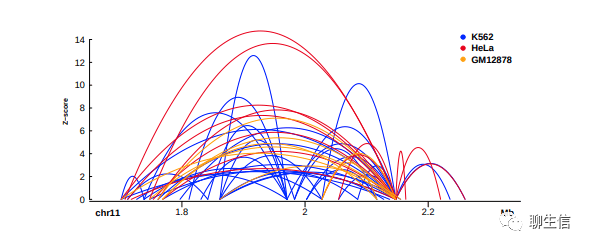
SparK
https://github.com/harbourlab/SparK
Publication: https://www.biorxiv.org/content/10.1101/845529v1.full.pdf
Language: Python
Tags: Static, Hi-C, Quantitative
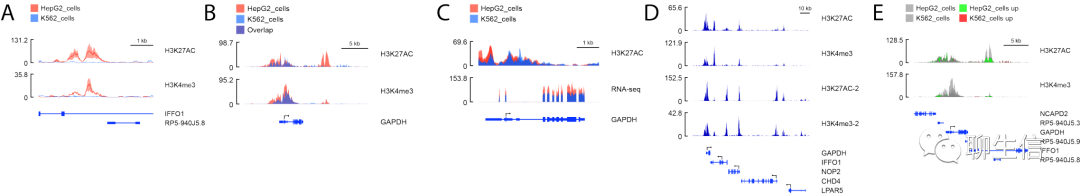
shinyChromosome
http://150.109.59.144:3838/shinyChromosome/
Publication: https://www.sciencedirect.com/science/article/pii/S1672022919301883
Language: R, Shiny
Tags: Dotplot, Linear, Ideogram
Platform: Web
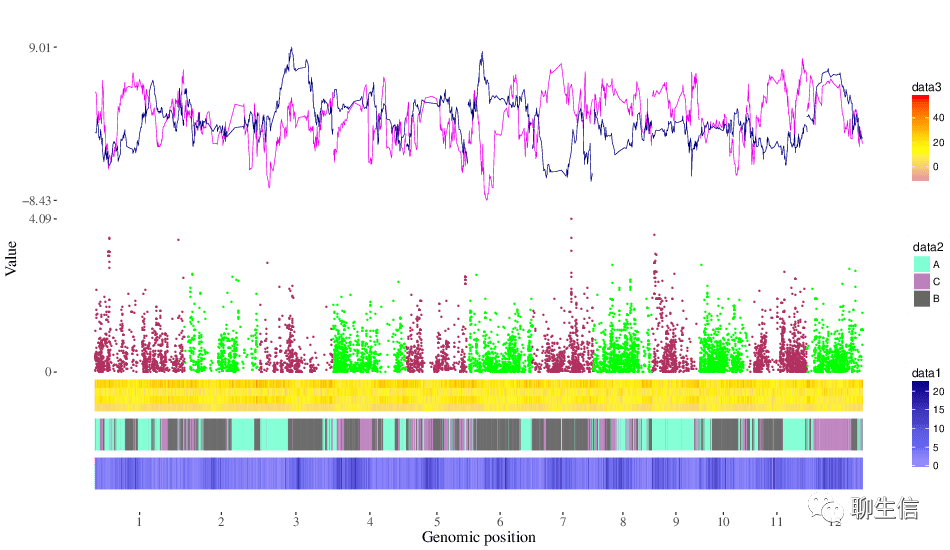
shabam
https://github.com/dlrice/shabam
Language: Python
Tags: Static
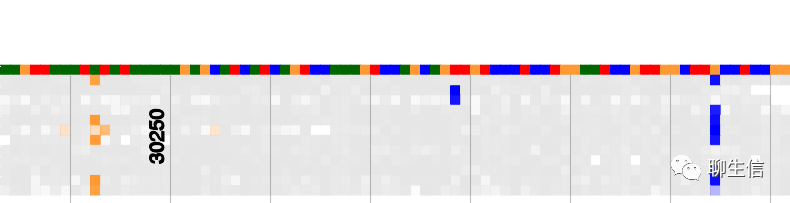
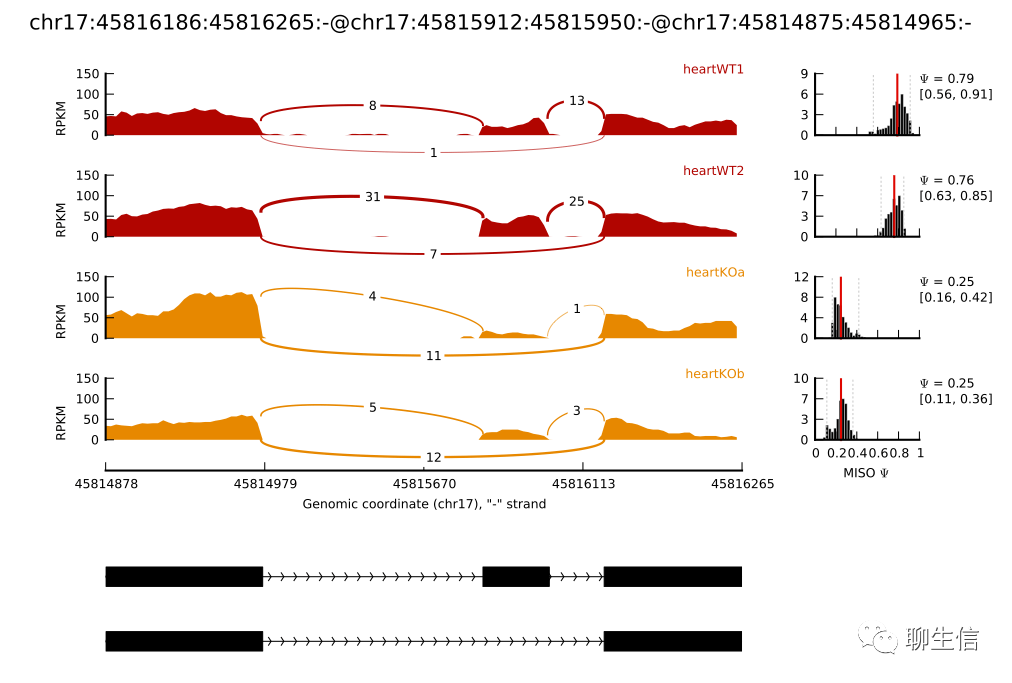
SashimiPlot
https://miso.readthedocs.io/en/fastmiso/sashimi.html
Language: Python
Tags: Static, Expression, Splicing
Samplot
https://github.com/ryanlayer/samplot
Language: Python
Tags: Static, SV, Alignment viewer
RepViz
https://bmcresnotes.biomedcentral.com/articles/10.1186/s13104-019-4473-z
Language: R
Tags: Static, Quantitative
Github: https://github.com/elolab/RepViz
RACER
https://oliviasabik.github.io/RACERweb/articles/IntroToRACER.html
Language: R
Tags: Static, GWAS
pyGenomeTracks
https://github.com/deeptools/pyGenomeTracks
Language: Python
Tags: Static, Hi-C
PureScript genetics browser
https://github.com/chfi/purescript-genetics-browser
Language: Purescript
Tags: GWAS
Platform: Web
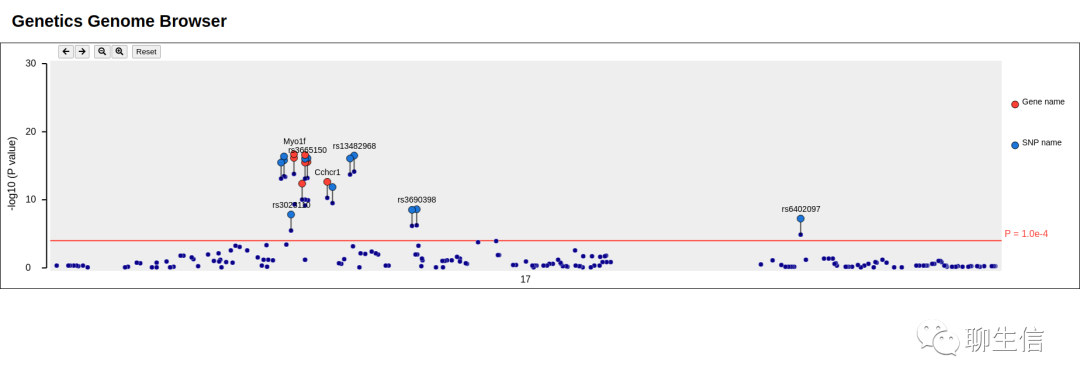
pauvre
https://github.com/conchoecia/pauvre
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6991124/figure/fig-3/
Language: Python
Tags: Static, Misc QC, Comparative
Platform: CLI
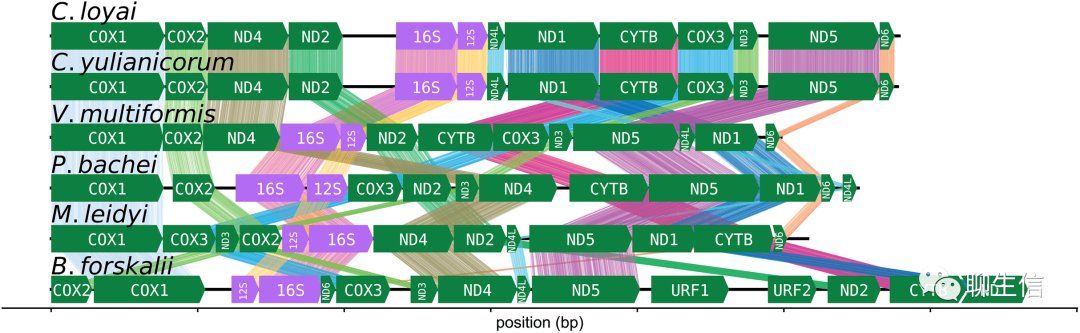
Pairoscope
http://pairoscope.sourceforge.net/
Language: C++
Tags: Static, SV
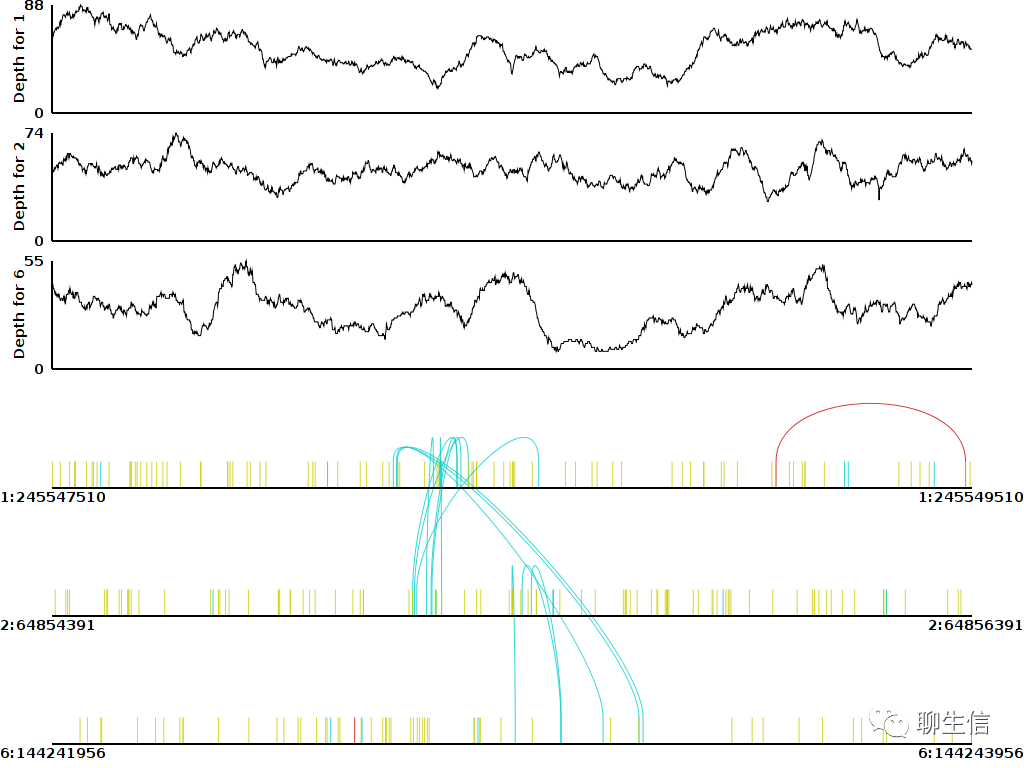
Millefy
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7053140/
Language: R
Tags: Single cell
Platform: Package
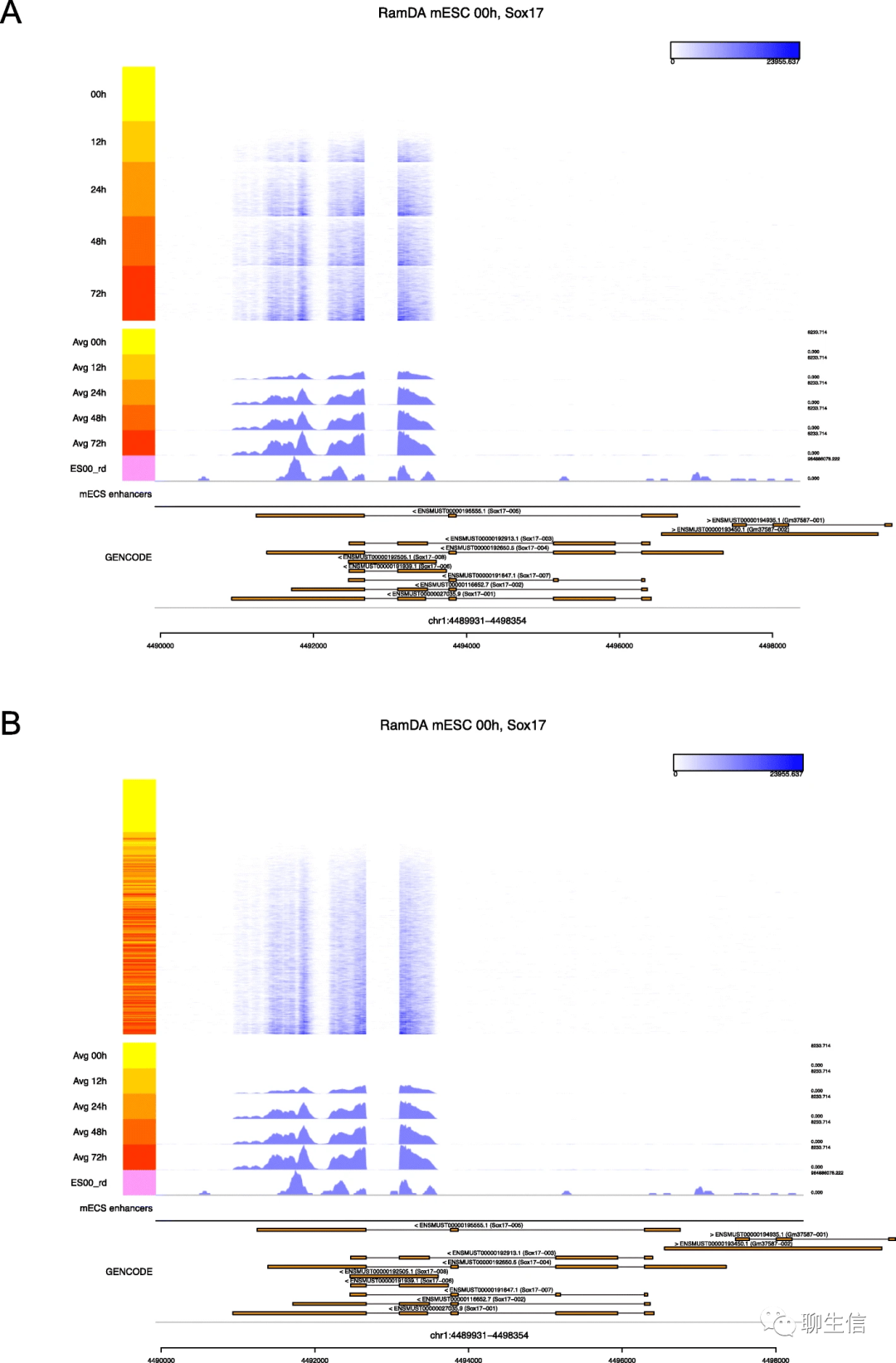
Methplotlib
https://www.biorxiv.org/content/10.1101/826107v1.full.pdf
Language: Python
Tags: Static, Epigenomics, Methylation
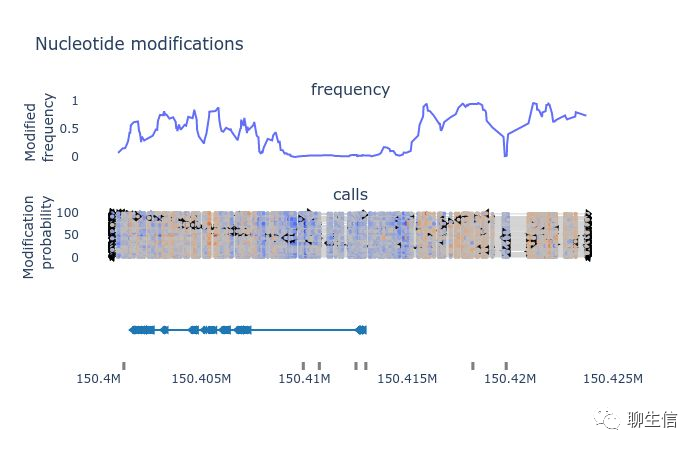
Mason
http://www.yeastrc.org/mason/
Tags: Static, Protein
Platform: Web
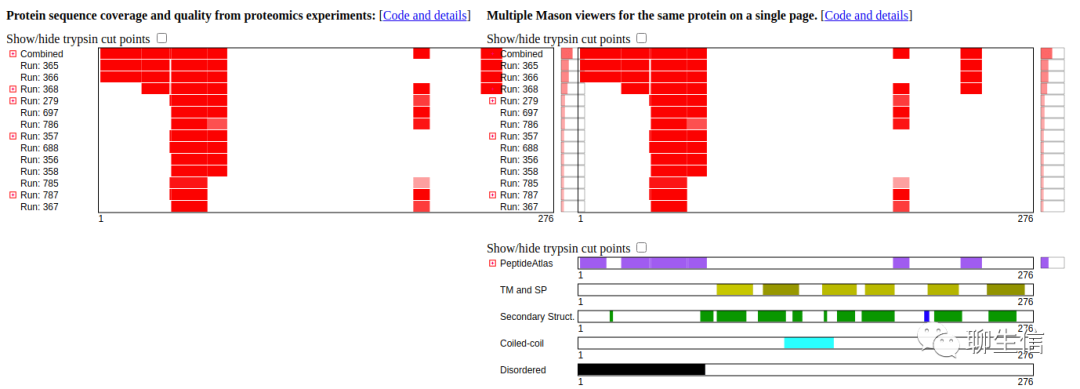
Lollipops
https://github.com/joiningdata/lollipops
Language: Go
Tags: Static, Protein, Lollipops
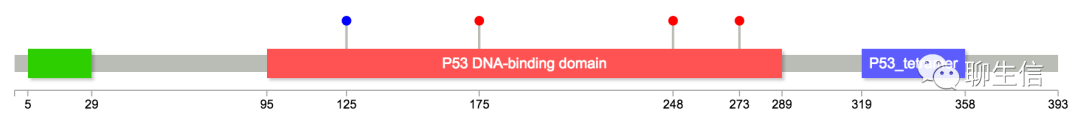
JVarKit/BamToSVG
http://lindenb.github.io/jvarkit/BamToSVG.html
Language: Java
Tags: Static, Alignment viewer

HiCPlotter
https://github.com/kcakdemir/HiCPlotter
Language: Python
Tags: Static, Hi-C
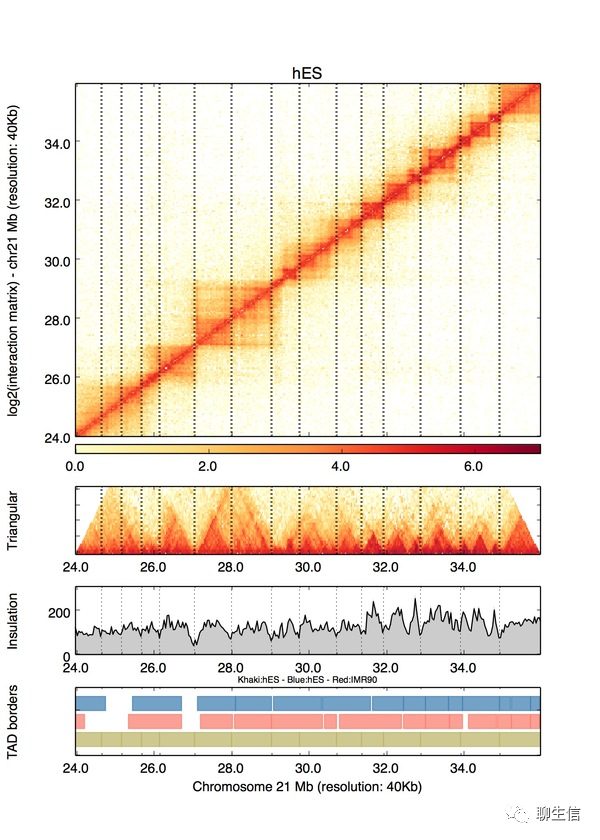
Hagfish
https://github.com/mfiers/hagfish/wiki/Plots
Language: Python
Tags: Static, Coverage
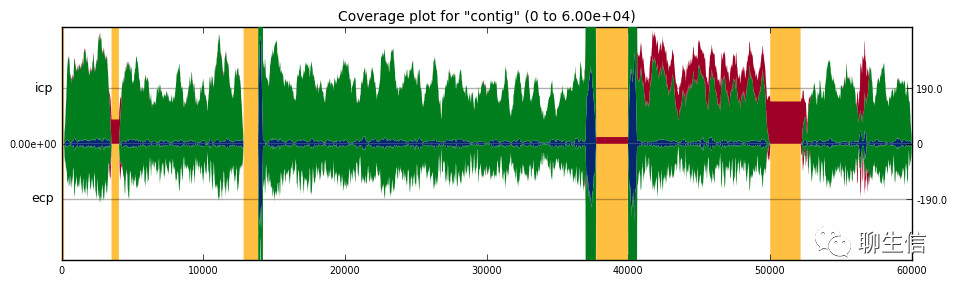
Gviz
https://bioconductor.org/packages/release/bioc/html/Gviz.html
Language: R
Tags: Static, Alignment viewer, Quantitative, Multi
gtrellis
http://bioconductor.org/packages/devel/bioc/vignettes/gtrellis/inst/doc/gtrellis.html
Language: R
Tags: Static, Ideogram, Quantitative, CNV
GSSPlayground
https://github.com/orangeSi/GSSplayground
Language: Perl
Tags: Comparative, Static, Alignment viewer

GGsashimi
https://github.com/guigolab/ggsashimi
Language: R
Tags: Static
GGgenes
https://github.com/wilkox/gggenes
Language: R
Tags: Static, Comparative
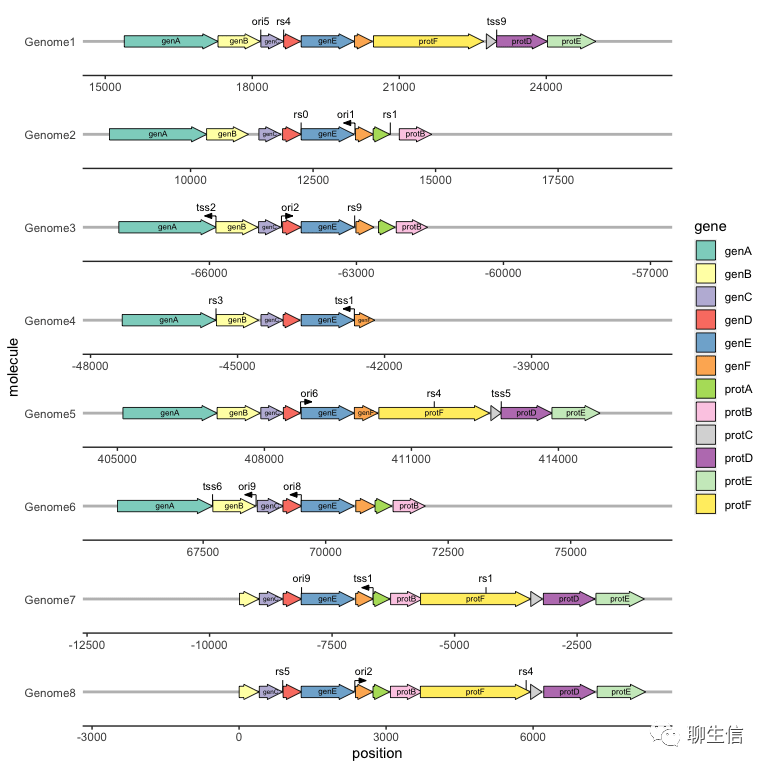
ggbio
http://www.bioconductor.org/packages/2.11/bioc/vignettes/ggbio/inst/doc/ggbio.pdf
Publication: https://genomebiology.biomedcentral.com/articles/10.1186/gb-2012-13-8-r77
Language: R
Tags: Static, Ideogram, Circular, Multi
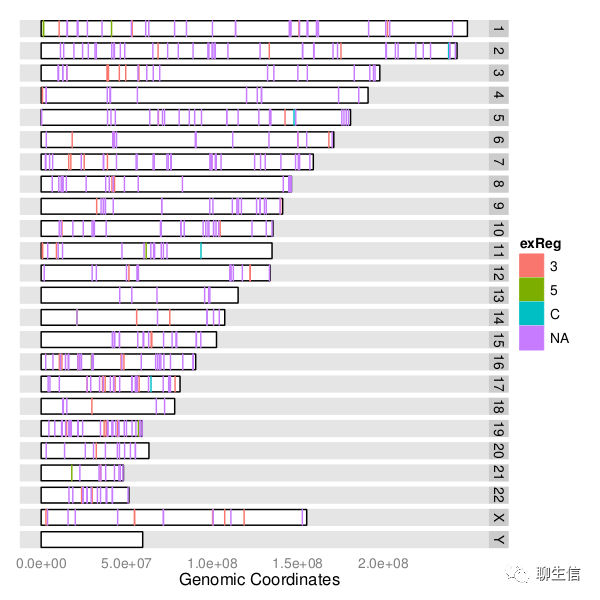
genoPlotR
http://genoplotr.r-forge.r-project.org/
Language: R
Tags: Static, Comparative
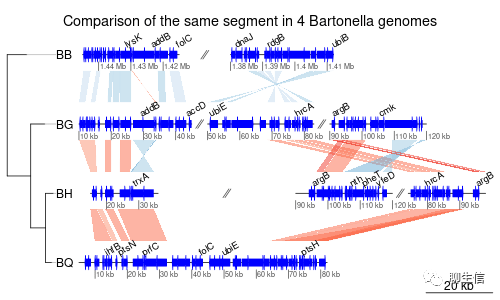
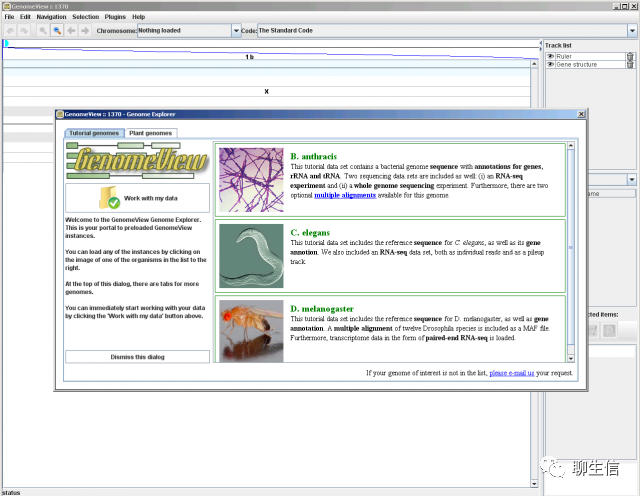
GenomeView (java app)
https://genomeview.org/content/quick-start-guide
Language: Java
Tags: Alignment viewer
GenomeView
https://github.com/nspies/genomeview
Language: Python
Tags: Static, SV, Alignment viewer
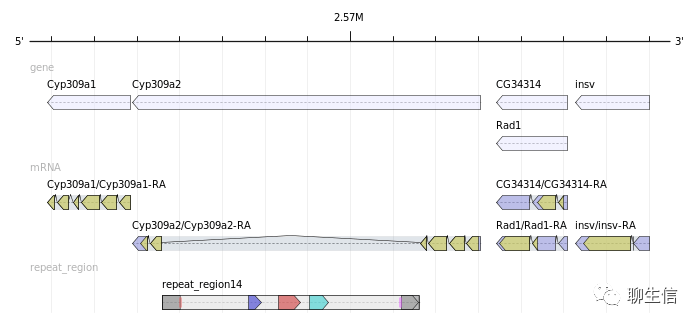
GenomeTools
http://genometools.org/
Language: C
Tags: Static, Gene structure
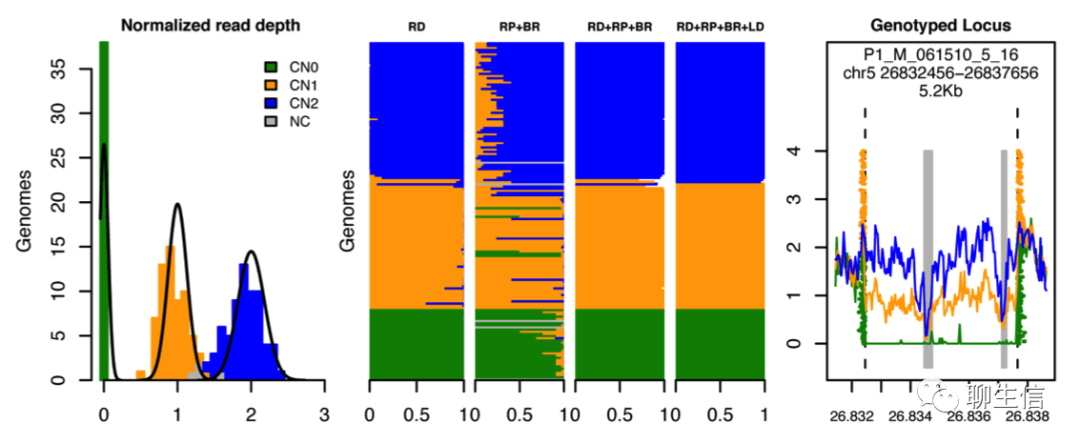
Genome STRiP
http://software.broadinstitute.org/software/genomestrip/
Tags: Static
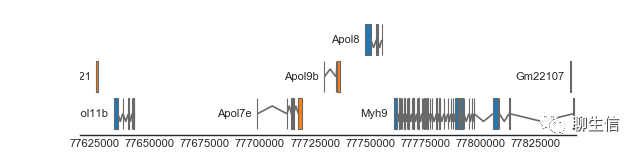
Geneviz
https://jrderuiter.github.io/geneviz/usage.html
Language: R
Tags: Static, Gene structure
FeatureViewer
https://github.com/calipho-sib/feature-viewer
Tags: Static, Gene structure
Platform: Web
ExonIntron
http://wormweb.org/exonintron
Tags: Static, Gene structure
DNAPlotLib
https://github.com/VoigtLab/dnaplotlib
Language: Python
Tags: Static, Synthetic biology
CNVpytor
https://github.com/abyzovlab/CNVpytor
Language: Python
Tags: Static, CNV
CNVPlot
https://github.com/dantaki/CNVplot/
Language: R
Tags: Static, CNV
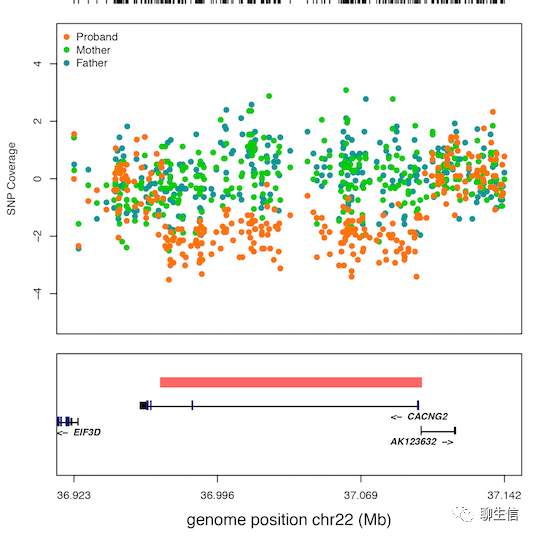
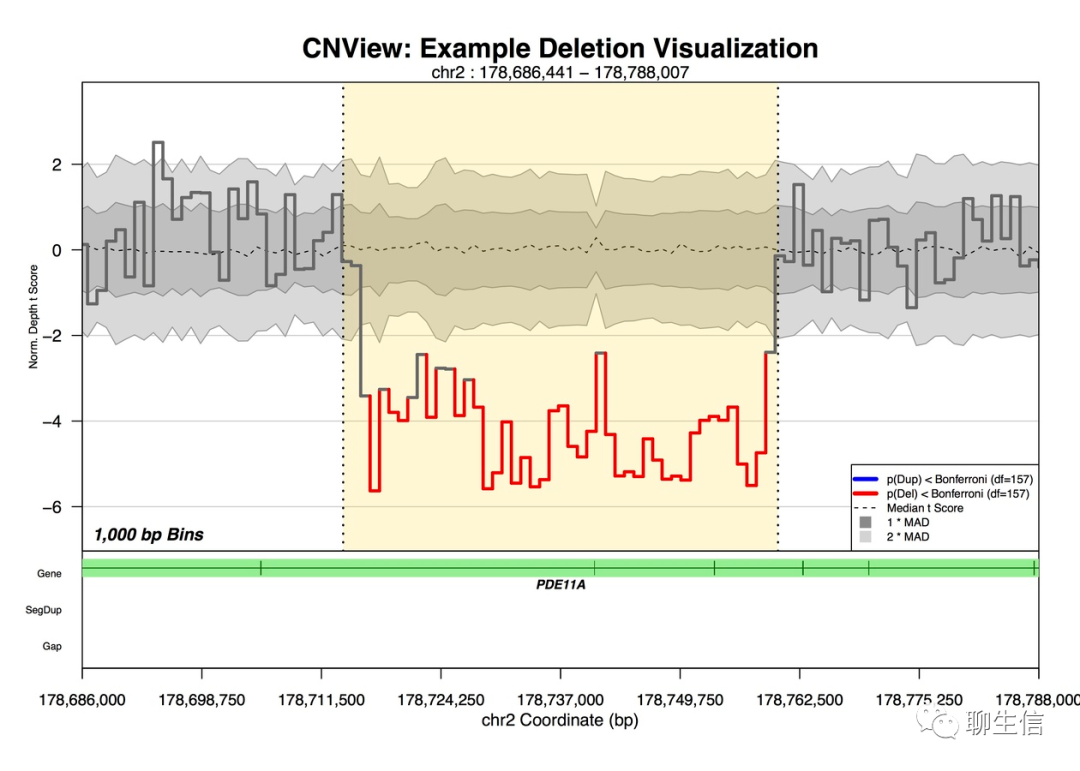
CNView
https://github.com/RCollins13/CNView
Language: R
Tags: Static, CNV
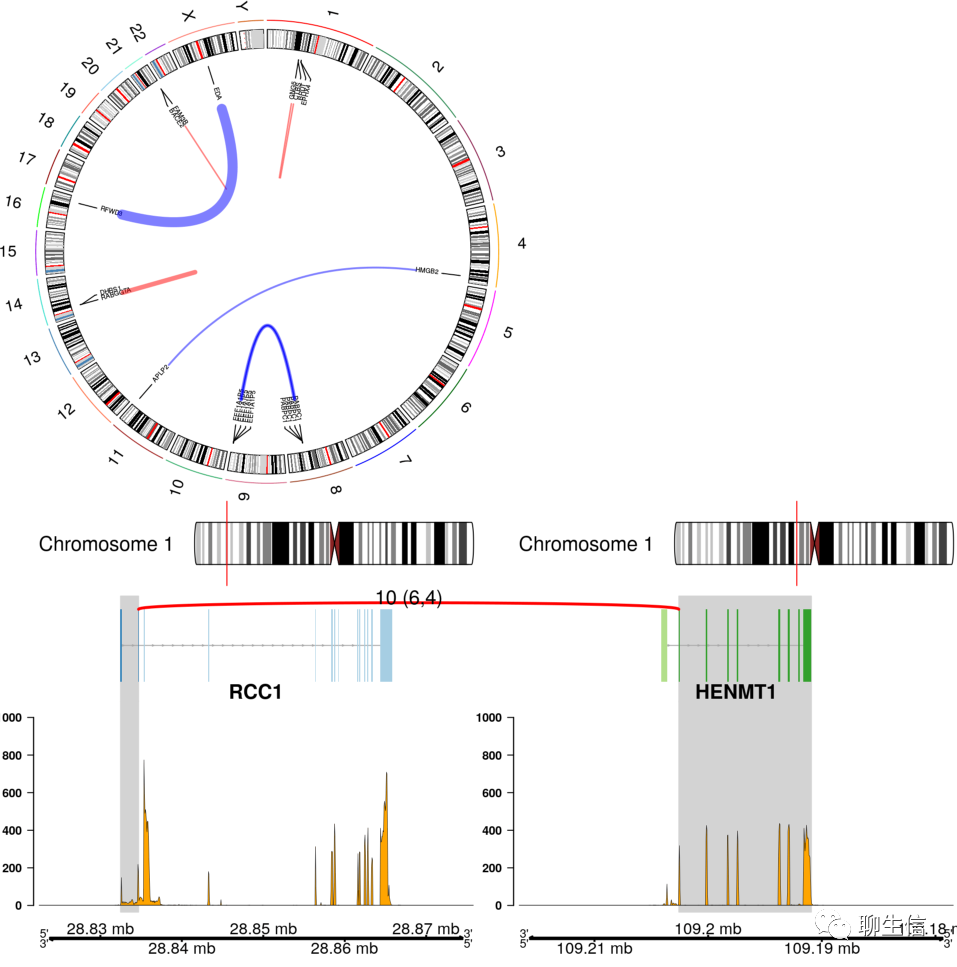
Chimeraviz
https://github.com/stianlagstad/chimeraviz
Language: R
Tags: Gene fusion, Circular, Linear
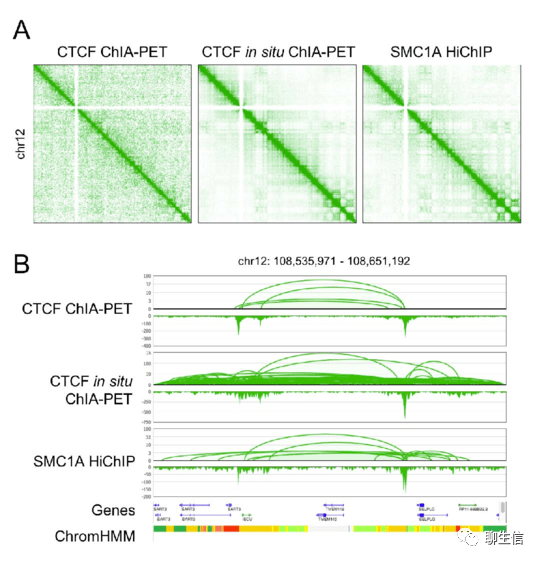
ChIA-Pipe
https://github.com/TheJacksonLaboratory/ChIA-PIPE
Publication: https://www.biorxiv.org/content/10.1101/506683v1.full.pdf
Tags: Static, Epigenomics
BAMSnap
https://github.com/parklab/bamsnap
Language: Python
Tags: Static, Alignment viewer
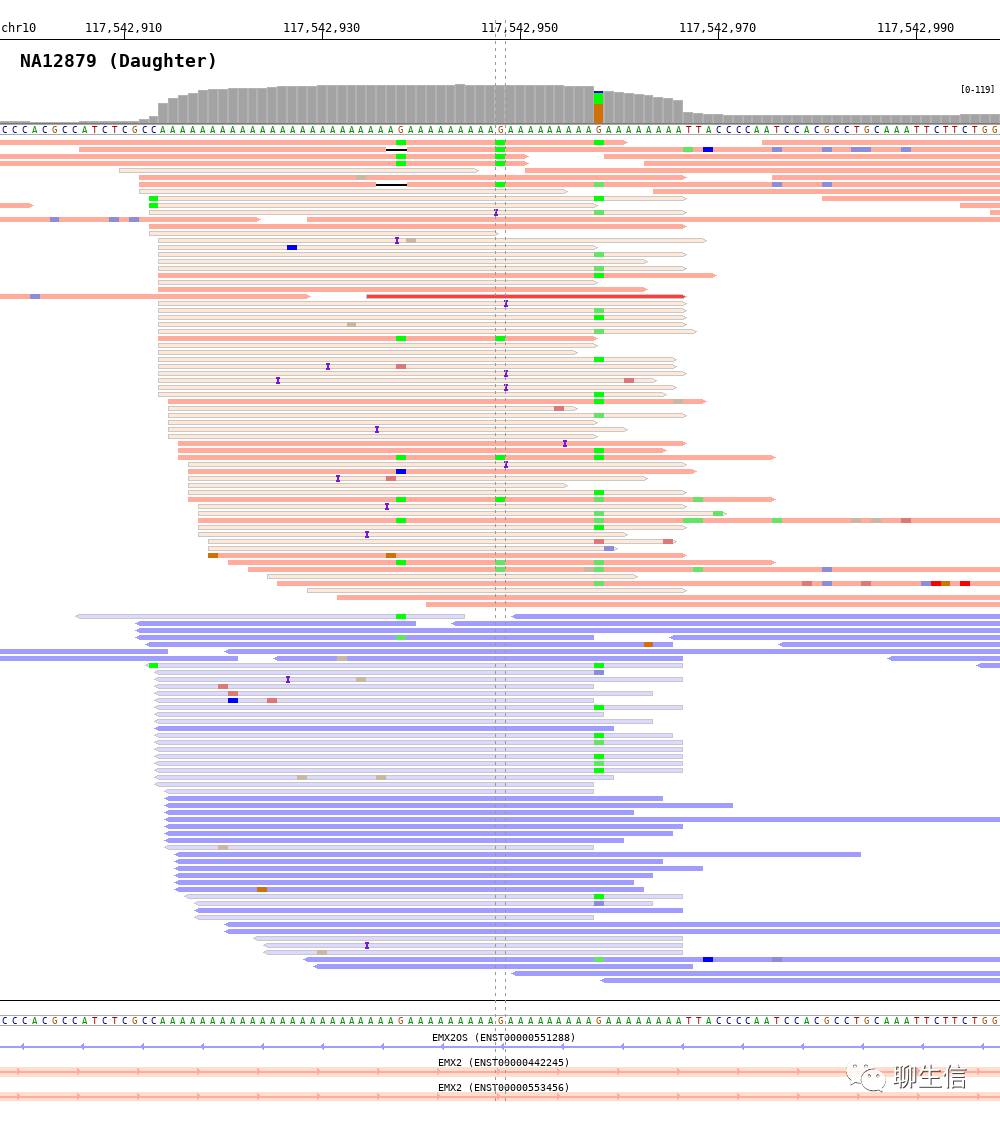
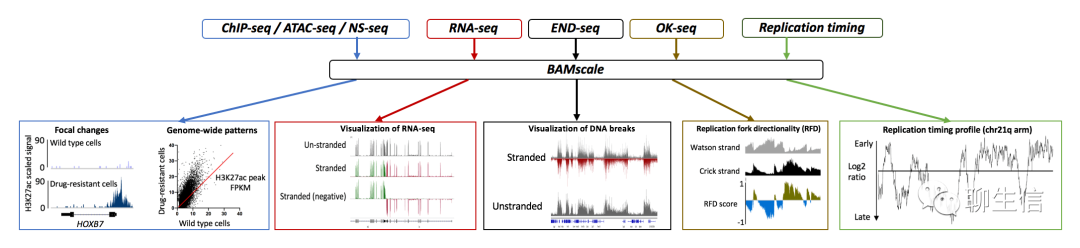
BAMScale
https://github.com/ncbi/BAMscale
Publication: https://epigeneticsandchromatin.biomedcentral.com/articles/10.1186/s13072-020-00343-x
Language: C, R
Tags: CNV, Static
CNVkit
https://cnvkit.readthedocs.io/en/stable/plots.html
Publication: https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1004873
Language: Python
Tags: CNV
Github: https://github.com/etal/cnvkit
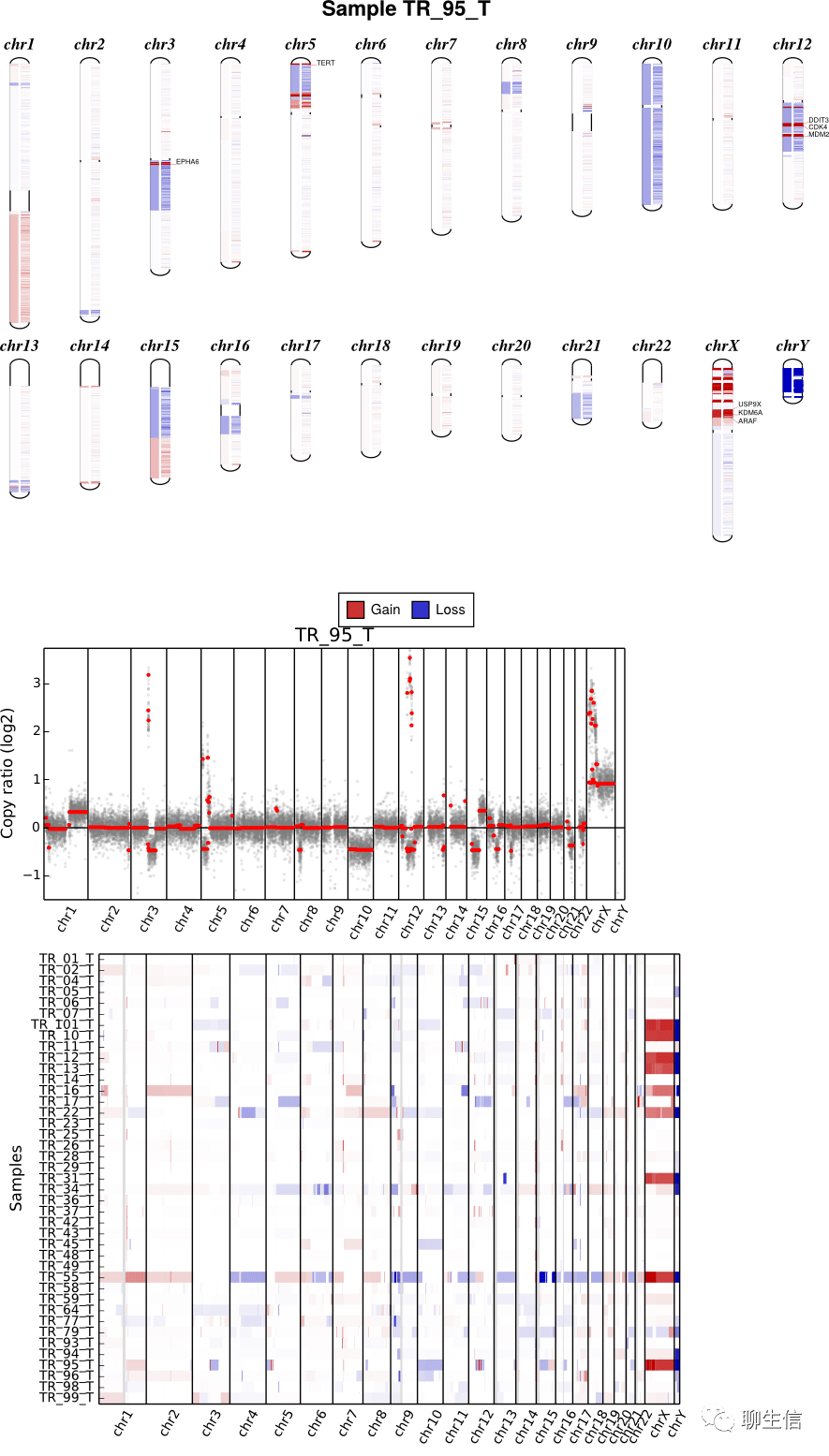
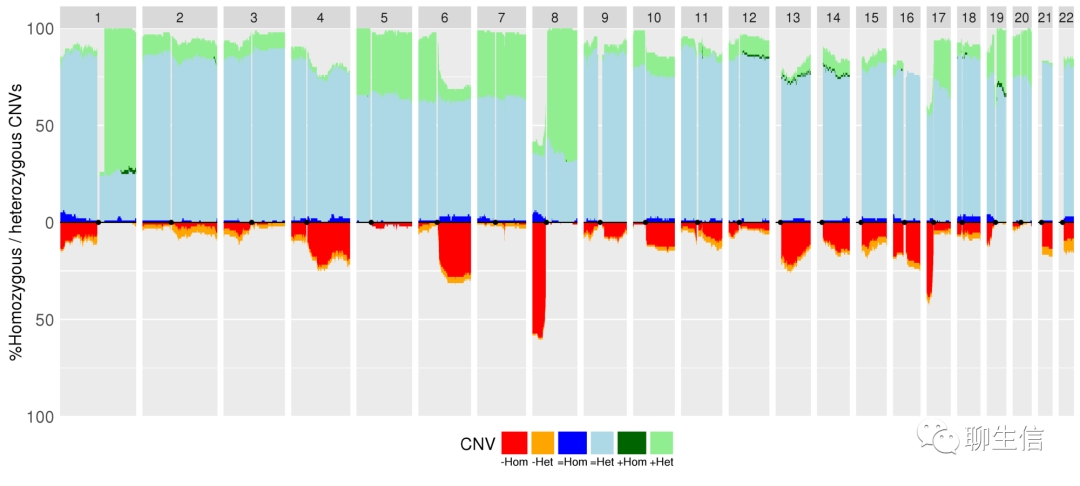
aCNVViewer
https://github.com/FJD-CEPH/aCNViewer
Publication: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0189334
Language: Python
Tags: CNV, Static
Wasabi
http://wasabiapp.org/
Tags: MSA
Platform: Web
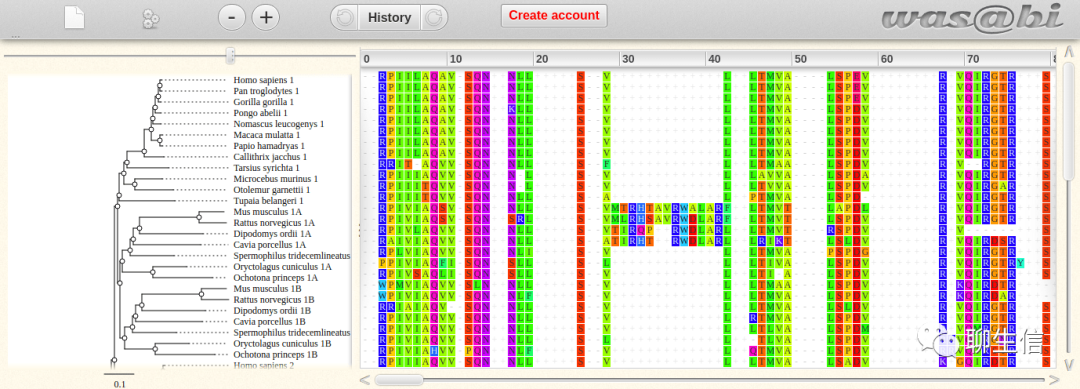
TeXShade
http://mirrors.ibiblio.org/CTAN/macros/latex/contrib/texshade/texshade.pdf
Tags: MSA
Platform: Desktop

STRAP
http://www.bioinformatics.org/strap/
Tags: MSA
Note: also see AA
Platform: Web, Applet

seqotron
https://github.com/4ment/seqotron
Language: Objective-C
Tags: MSA
Platform: Desktop
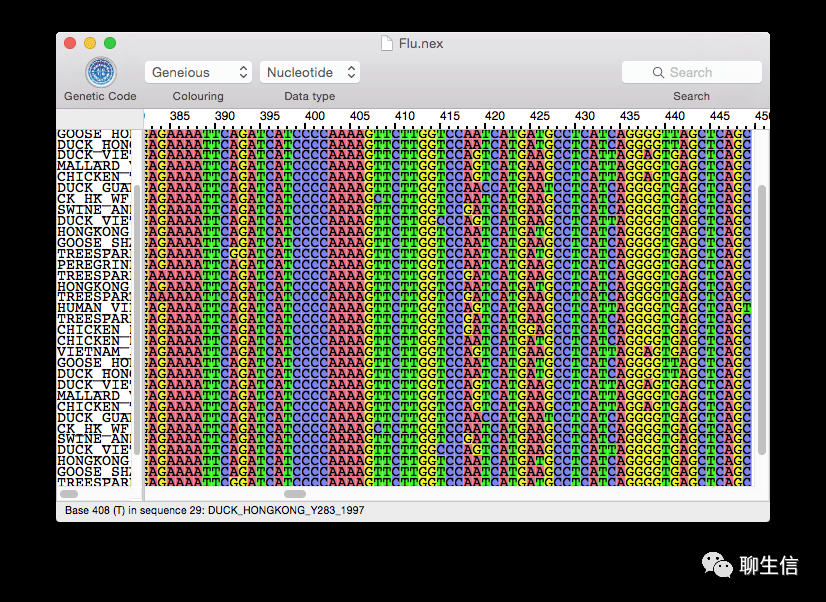
react-msaview
https://github.com/GMOD/react-msaview
Language: JS, React
Tags: MSA
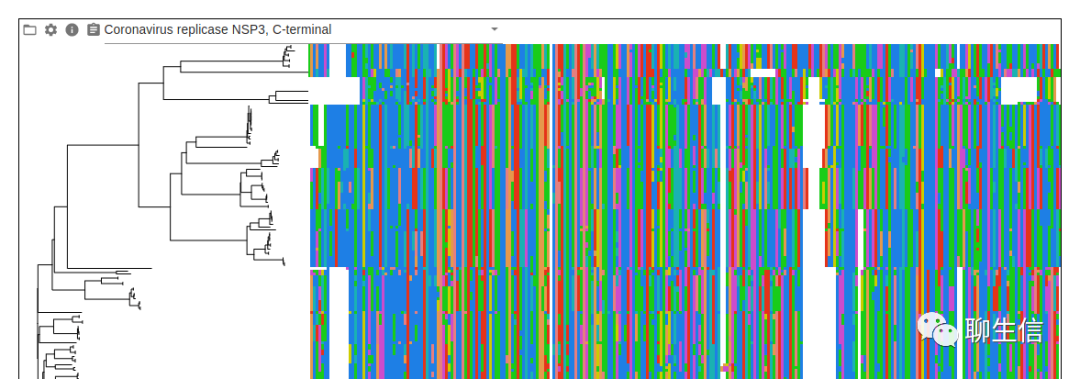
React MSAViewer
https://github.com/plotly/react-msa-viewer
Language: JS, React
Tags: MSA
Platform: Web
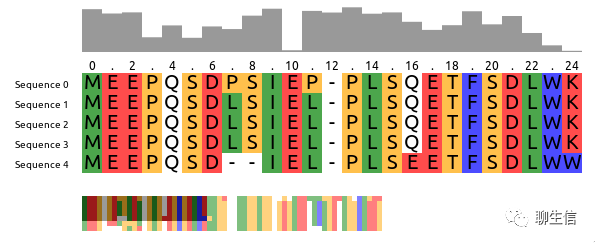
pymsaplotter
https://github.com/orangeSi/pymsaploter
Language: Python
Tags: MSA
pyBoxshade
https://github.com/mdbaron42/pyBoxshade
Language: Python
Tags: MSA
Note: also see boxshade
Platform: Desktop
NCBI MSA Viewer
https://www.ncbi.nlm.nih.gov/projects/msaviewer/
Tags: MSA
Platform: Web, CGI
MView
https://www.ebi.ac.uk/Tools/msa/mview/
Tags: MSA
Platform: Web
msaR
https://github.com/zachcp/msaR
Language: R, JS
Tags: MSA
Note: uses BioJS/MSA
Platform: Desktop, R/htmlwidget
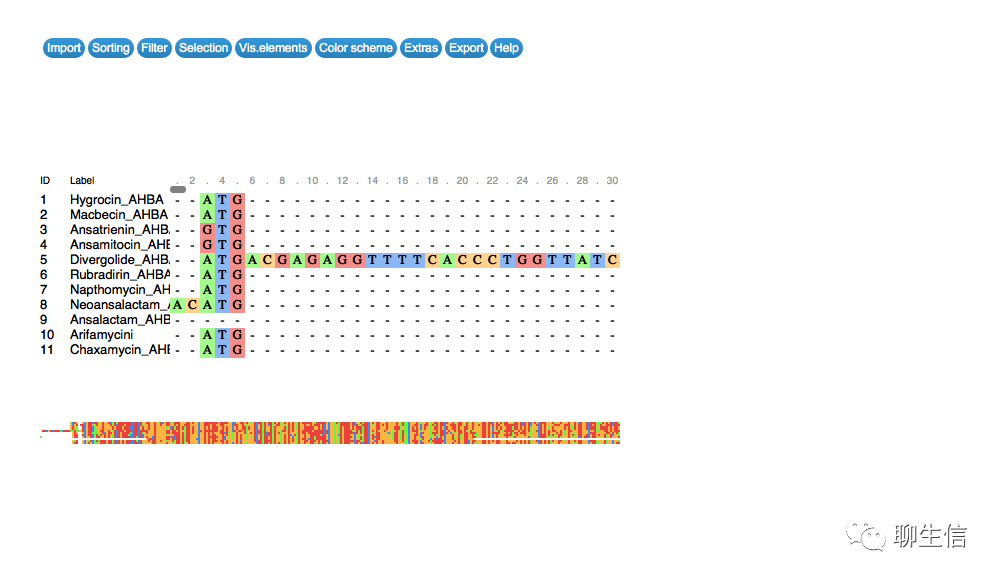
MSABrowser
https://thekaplanlab.github.io/
Tags: MSA
Platform: Web
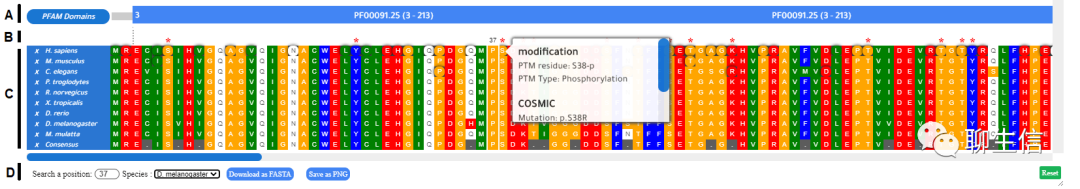
LogoJS
https://logojs.wenglab.org/app/gallery/
Language: JS
Tags: Logo
Platform: Web
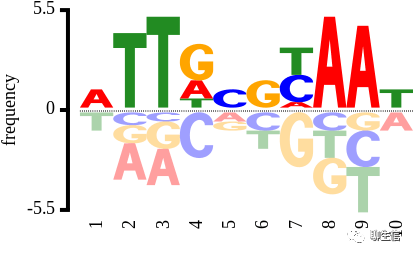
JSAV
http://www.bioinf.org.uk/software/jsav
Publication: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4304231/
Language: JS
Tags: MSA
Platform: Web
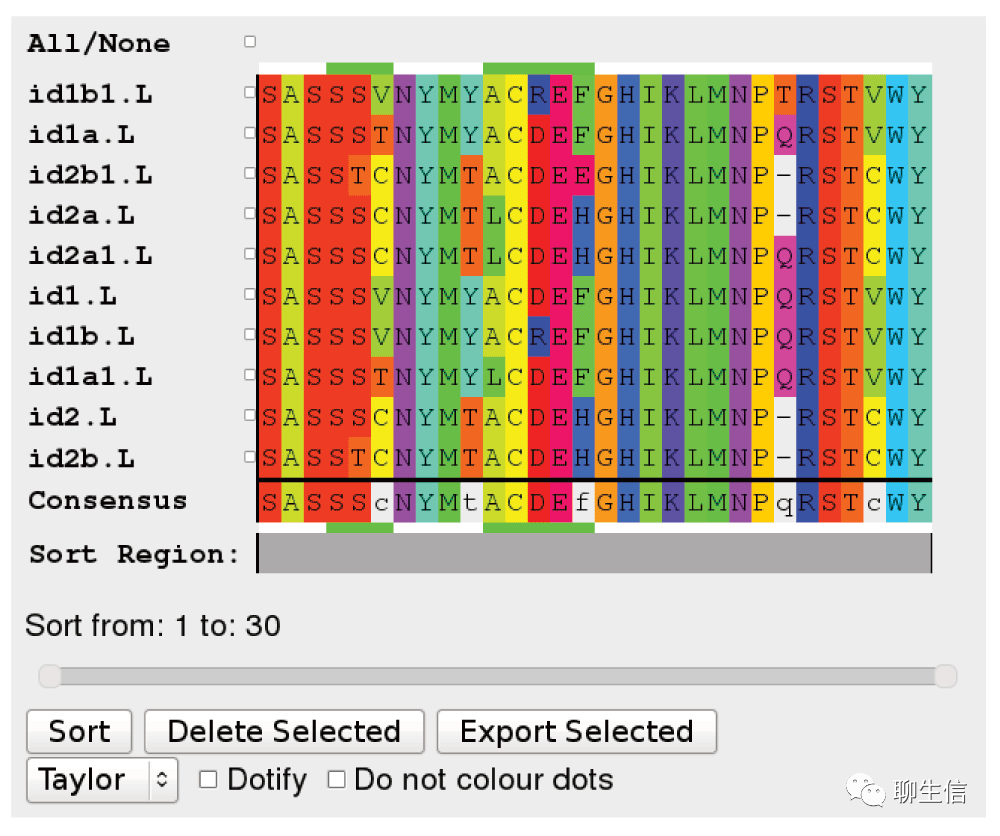
Jalview
https://www.jalview.org/
Language: Java
Tags: MSA
Platform: Desktop
ESPript
http://espript.ibcp.fr/ESPript/ESPript/
Tags: MSA
Platform: Desktop
Boxshade
https://embnet.vital-it.ch/software/BOX_form.html
Tags: MSA
Platform: Web

BioJS/MSA
https://msa.biojs.net/
Publication: https://academic.oup.com/bioinformatics/article/32/22/3501/2525598?login=true
Tags: MSA
Platform: Web
AliView
http://www.ormbunkar.se/aliview/
Language: Java
Tags: MSA
Platform: Desktop
AlignmentViewer
https://alignmentviewer.org/
Tags: MSA
Platform: Web
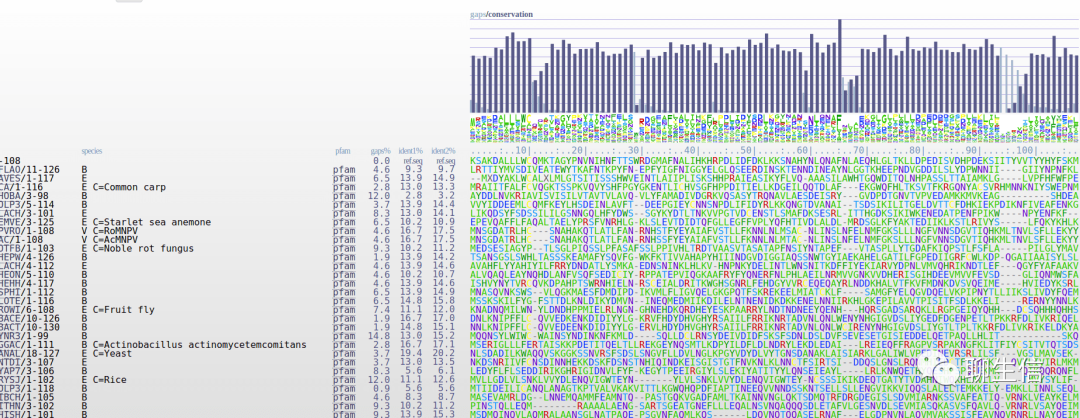
AlignmentComparator
http://bioinfweb.info/AlignmentComparator/
Language: Java
Tags: MSA
Note: Uses http://bioinfweb.info/LibrAlign/
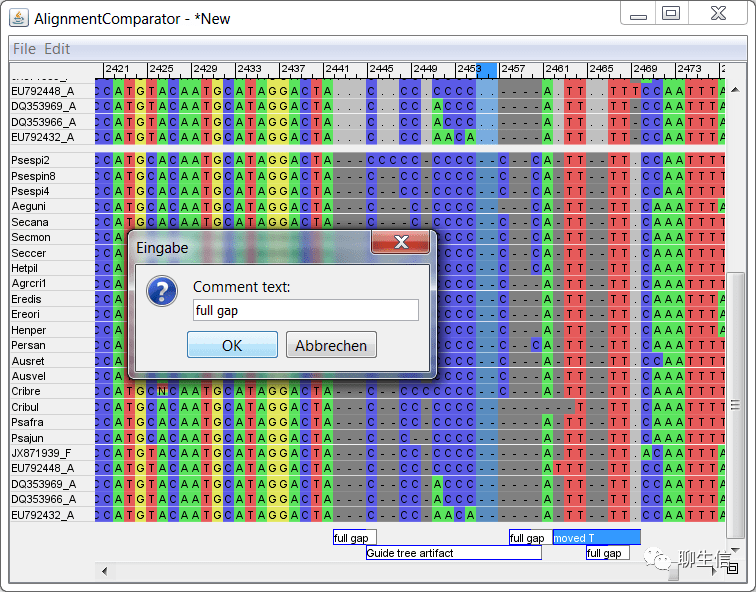
alignment.js
https://github.com/veg/alignment.js
Tags: MSA
Platform: Web
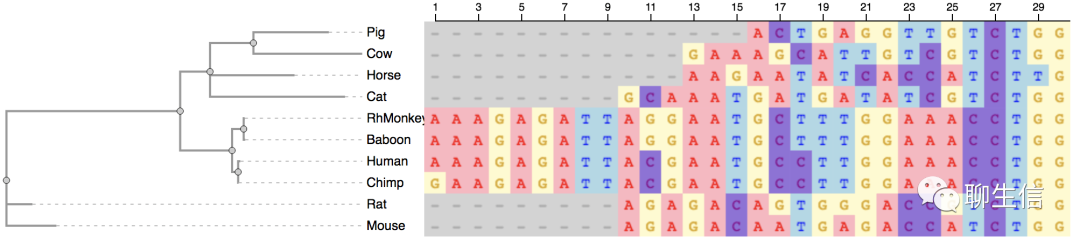
abrowse
https://github.com/ihh/abrowse
Tags: MSA
Platform: Web
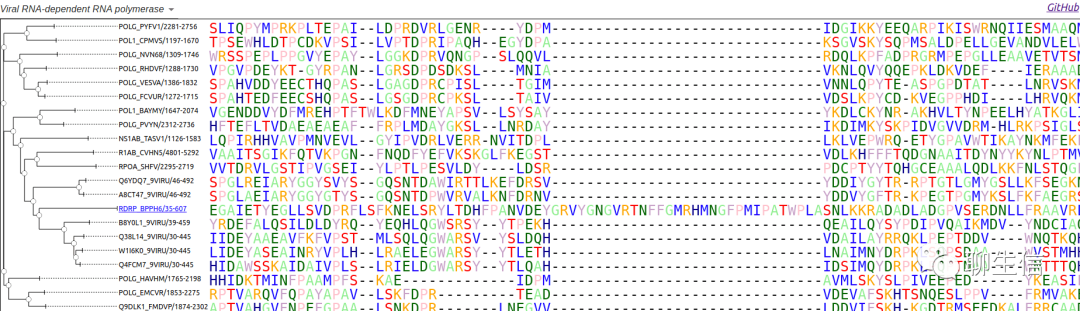
AA
http://www.bioinformatics.org/strap/aa
Tags: MSA
Note: also see STRAP
Platform: Web
Bionano
https://bionanogenomics.com/technology/structural-variation/
Tags: Commercial, SV
Strand NGS
https://www.strand-ngs.com/
Tags: Commercial
SnapGene
https://www.snapgene.com/
Tags: Commercial
Persephone
https://persephonesoft.com/
Tags: Commercial, Comparative
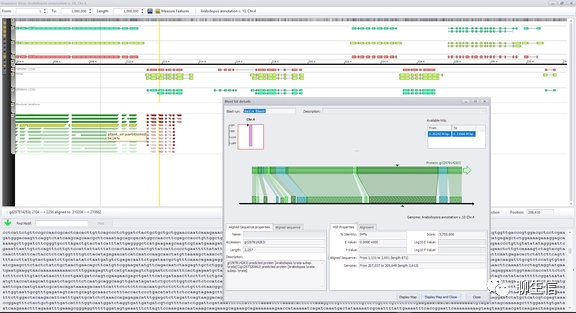
MacVector
https://macvector.com/
Tags: Commercial
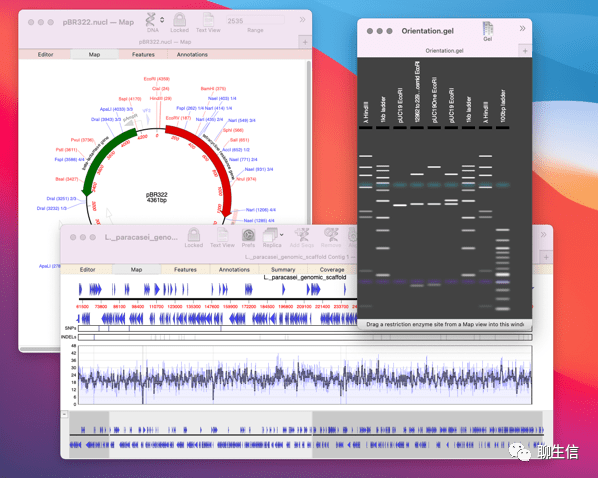
Lucid viewer
https://lucidalign.com/
Tags: Commercial
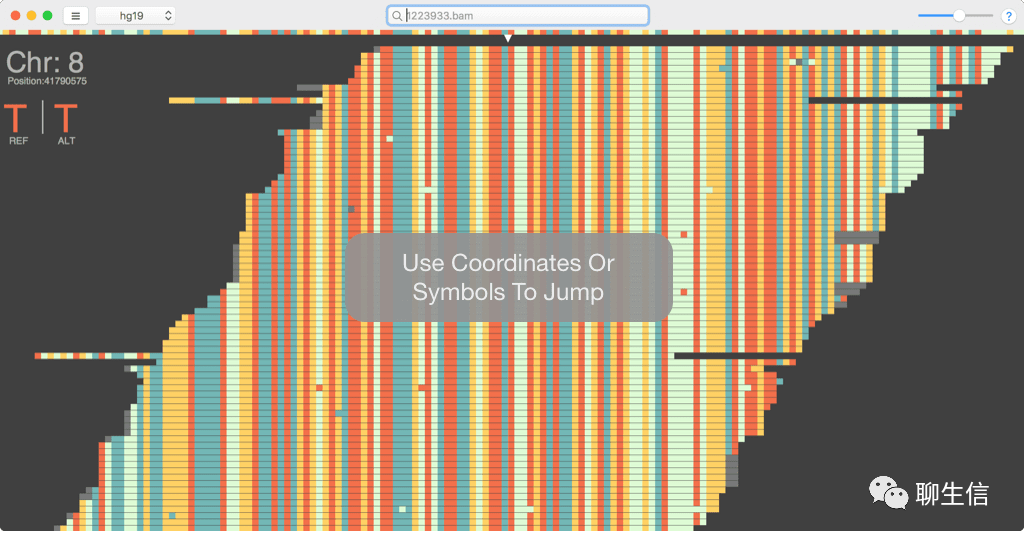
Golden Helix
http://goldenhelix.com/GenomeBrowse/
Tags: Commercial
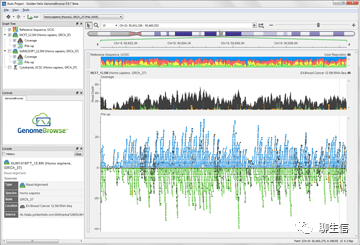
Genestack
https://genestack.com/blog/2015/05/28/navigation-in-genestack-genome-browser/
Tags: Commercial
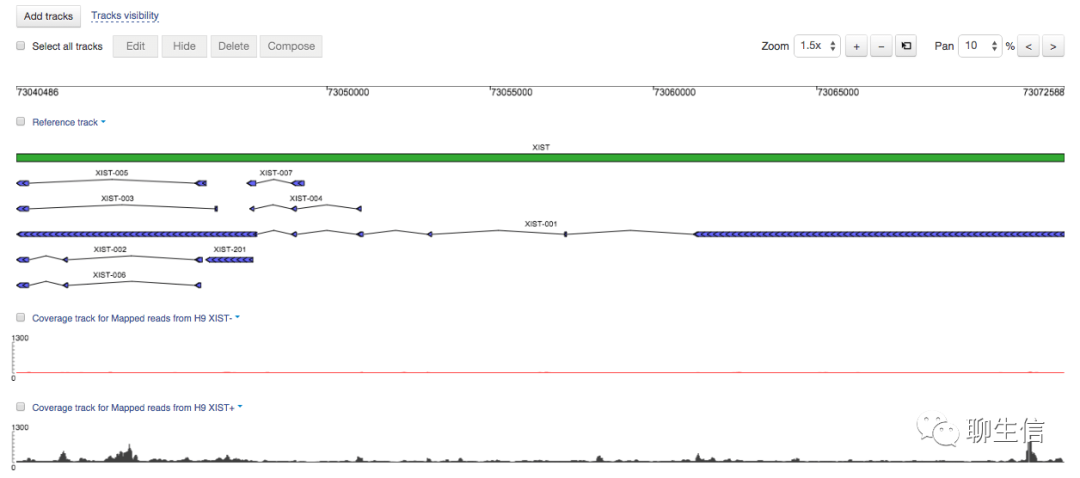
Geneious
https://www.geneious.com/
Tags: Commercial, Synthetic biology
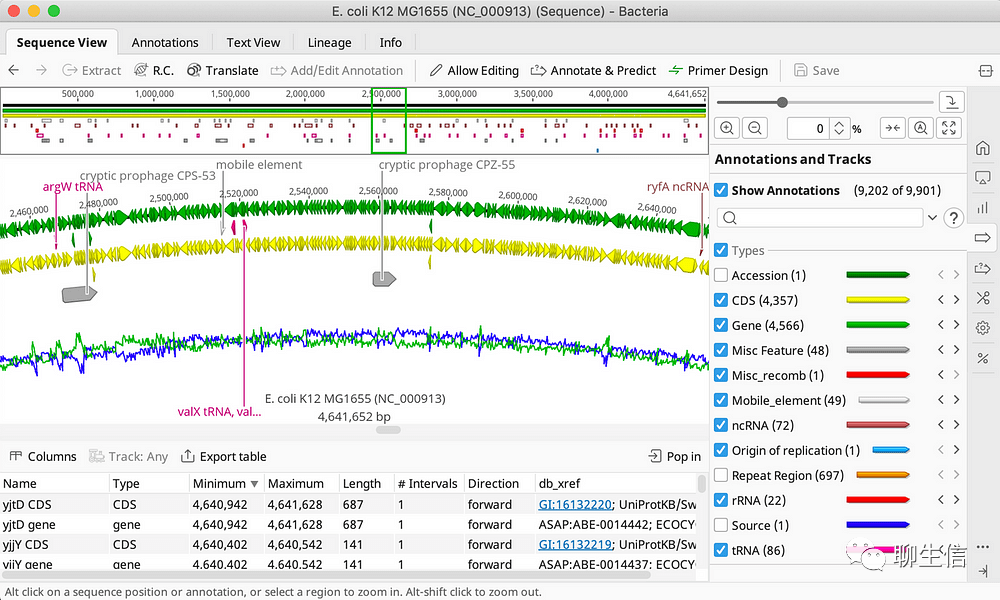
DNASTAR
https://www.dnastar.com/
Tags: Commercial
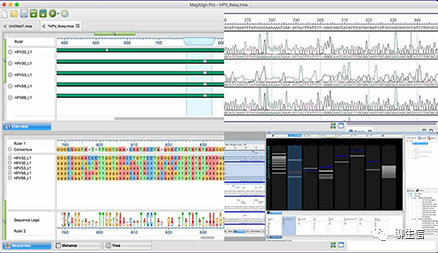
CLC Genomics workbench
https://digitalinsights.qiagen.com/news/blog/discovery/structural-variant-detection-using-clc-genomics-workbench/
Tags: Commercial, SV
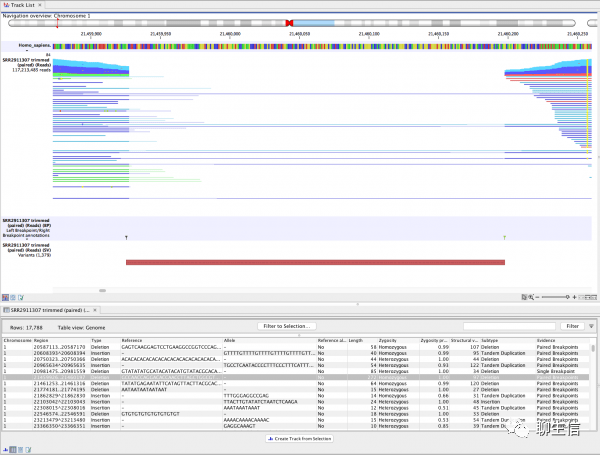
Benchling
https://www.benchling.com/
Tags: Commercial, Synthetic biology
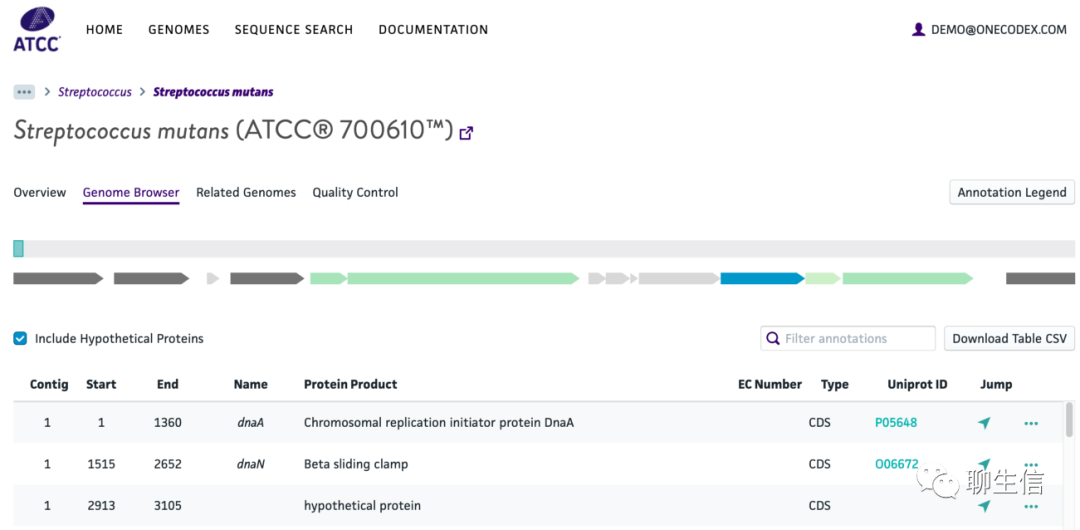
ATCC Genome Portal (American Type Culture Collection)
https://docs.onecodex.com/en/articles/3996697-using-the-genome-browser
Tags: Commercial, Microbiology
Alamut
https://www.interactive-biosoftware.com/alamut-visual/
Tags: Commercial
10x genomics/Loupe
https://support.10xgenomics.com/genome-exome/software/visualization/latest/structural-variants
Tags: Commercial

Note: if you would like your tool removed or screenshot removed (for copyright purposes for example) let me know


