图神经网络预训练 (3) - Context Prediction + 监督学习 代码
赞
踩
前两篇内容概述了Weihua Hu*, Bowen Liu*图神经网络预训练的方法,以及context prediction进行预训练的实施代码。
context prediction 学习的图内的原子/边信息的表征,并没有包括图层面的信息。
这一部分的监督学习,是图层次的监督学习,目的是把图层面的信息增加到图的表征向量G(h)中。经过图层次的监督学习,得到的模型就可以直接用于下游的任务。
文章方法:在节点层面预训练的模型后加上一个简单的线性模型,用于图层面的监督训练
网络结构如下图:
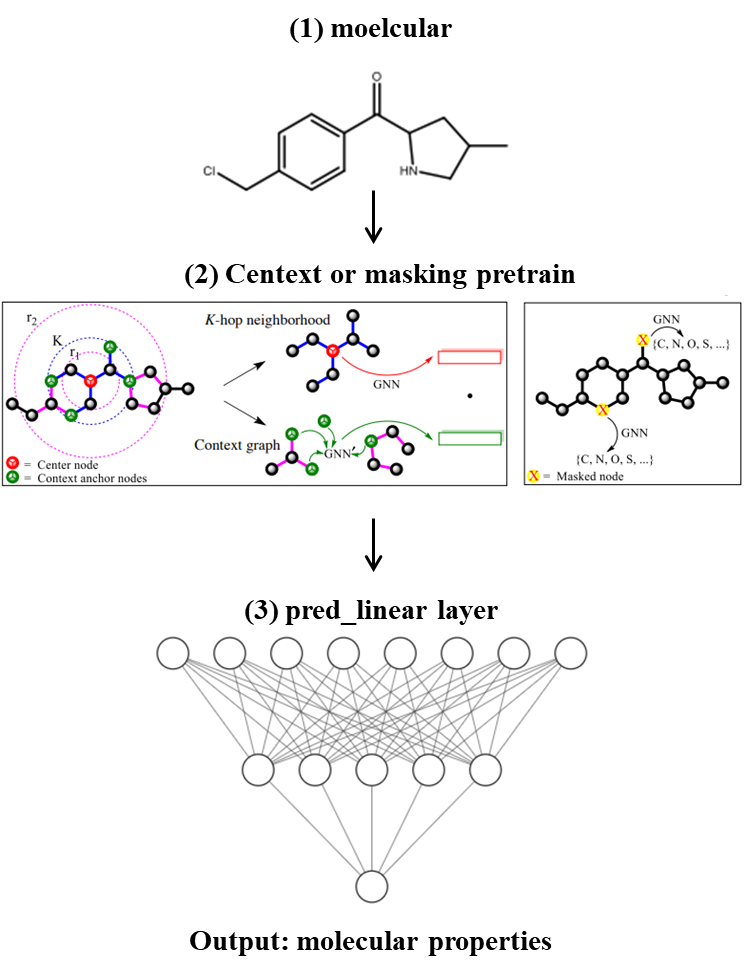
在文献中,作者的图层面任务的监督学习是多任务学习的方法,使用chembl_filtered数据集。再经过这一层训练以后,往往还加上Fine-tuning,也就是特定任务的训练,例如:BBBP。
但是由于版本问题,chembl_filtered数据集无法加载。所以这里使用esol和lipophilicity等数据集,直接作为Supervised pre-training和Fine-tuning。
以下为代码部分:
一、导入相关包
- 导入相关包
- import pandas as pd
- from tqdm import tqdm
- import numpy as np
-
- import os
- import math
- import random
- import torch
- import torch.optim as optim
- torch.manual_seed(0)
- np.random.seed(0)
-
- from rdkit import Chem
- from rdkit.Chem import Descriptors
- from rdkit.Chem import AllChem
- from rdkit import DataStructs
- from rdkit.Chem.rdMolDescriptors import GetMorganFingerprintAsBitVect
-
- import torch.nn.functional as F
- from torch_geometric.data import Data
- from torch_geometric.data import DataLoader
- from torch_geometric.data import InMemoryDataset
- from torch_geometric.nn import MessagePassing, global_add_pool, global_mean_pool, global_max_pool, GlobalAttention, Set2Set
-
- from torch_geometric.utils import add_self_loops, degree, softmax
- from torch_geometric.nn.inits import glorot, zeros
-
- from sklearn.model_selection import train_test_split
- import seaborn as sns
- import matplotlib.pyplot as plt
-
- #运行设备
- device = torch.device(torch.device('cuda')if torch.cuda.is_available() else torch.device('cpu'))

数据加载,大部分类似与预训练的时候,但是有标签,放在data.y里面。
data.y = torch.tensor([label])分子预处理过程和结果与预训练时一致。这里也是定义一个MolecularDataset。读取文件的方式上,有一些变化,因为监督学习的数据格式是csv:
- input_path = self.raw_paths[0]
- input_df = pd.read_csv(input_path, sep=',', dtype='str')
-
- smiles_list = list(input_df['smiles'])
- smiles_id_list = list(input_df.index.values)
- y_list = list(input_df['exp'].values)
数据加载部分代码:
-
-
- #PYG数据集
- class MoleculeDataset(InMemoryDataset):
- '''
- 将zinc数据集加载成PYG的Dataset
- '''
-
- def __init__(self, root, dataset='zinc250k',
- transform=None, pre_transform=None,
- pre_filter=None):
-
- self.dataset = dataset
-
- self.root = root
- super(MoleculeDataset, self).__init__(root, transform, pre_transform,
- pre_filter) # 要放在后面
- print(self.processed_paths[0])
- self.data, self.slices = torch.load(self.processed_paths[0])
-
- @property # 返回原始文件列表
- def raw_file_names(self):
- file_name_list = os.listdir(self.raw_dir)
- return file_name_list
-
- @property # 返回需要跳过的文件列表
- def processed_file_names(self):
- return 'geometric_data_processed.pt'
-
- def process(self):
- data_smiles_list = []
- data_list = []
-
- input_path = self.raw_paths[0]
- input_df = pd.read_csv(input_path, sep=',', dtype='str')
-
- smiles_list = list(input_df['smiles'])
- smiles_id_list = list(input_df.index.values)
- y_list = list(input_df['exp'].values)
-
- for i in range(len(smiles_list)):
- if i % 1000 == 0:
- print(str(i) + '...')
- s = smiles_list[i]
- label = float(y_list[i])
- # each example contains a single species
- try:
- rdkit_mol = AllChem.MolFromSmiles(s, sanitize=True)
- if rdkit_mol != None: # ignore invalid mol objects
- # # convert aromatic bonds to double bonds
- # Chem.SanitizeMol(rdkit_mol,sanitizeOps=Chem.SanitizeFlags.SANITIZE_KEKULIZE)
-
- data = mol_to_graph_data_obj_simple(rdkit_mol)
- if 119 in list(data.x[:, 0]):
- print(s)
- if 4 in list(data.edge_attr[:, 0]):
- print(s)
- # manually add mol id
- id = int(smiles_id_list[i])
- data.id = torch.tensor([id])
- data.y = torch.tensor([label])
- # data.y = torch.tensor([y_list[i]])
- # print('NNNNN')
- # print(y_list)
- data_list.append(data)
- data_smiles_list.append(smiles_list[i])
-
- except:
- continue
-
- # 过滤器
- if self.pre_filter is not None:
- data_list = [data for data in data_list if self.pre_filter(data)]
-
- # 转换器,
- if self.pre_transform is not None:
- data_list = [self.pre_transform(data) for data in data_list]
-
- # write data_smiles_list in processed paths
- data_smiles_series = pd.Series(data_smiles_list)
- data_smiles_series.to_csv(os.path.join(self.processed_dir,
- 'smiles.csv'), index=False,
- header=False)
- # InMemoryDataset的方法,将 torch_geometric.data.Data的list,转化为内部存储
- # 这里设置的保存路径为processedpath[0]
- data, slices = self.collate(data_list)
- torch.save((data, slices), self.processed_paths[0])
-
- # 显示属性
- def __repr__(self):
- return '{}()'.format(self.dataname)
-

- #从SMILES生成PYG的数据类型,与预训练过程一致
- allowable_features = {
- 'possible_atomic_num_list': list(range(1, 119)),
- 'possible_formal_charge_list': [-5, -4, -3, -2, -1, 0, 1, 2, 3, 4, 5],
- 'possible_chirality_list': [
- Chem.rdchem.ChiralType.CHI_UNSPECIFIED,
- Chem.rdchem.ChiralType.CHI_TETRAHEDRAL_CW,
- Chem.rdchem.ChiralType.CHI_TETRAHEDRAL_CCW,
- Chem.rdchem.ChiralType.CHI_OTHER
- ],
- 'possible_hybridization_list': [
- Chem.rdchem.HybridizationType.S,
- Chem.rdchem.HybridizationType.SP, Chem.rdchem.HybridizationType.SP2,
- Chem.rdchem.HybridizationType.SP3, Chem.rdchem.HybridizationType.SP3D,
- Chem.rdchem.HybridizationType.SP3D2, Chem.rdchem.HybridizationType.UNSPECIFIED
- ],
- 'possible_numH_list': [0, 1, 2, 3, 4, 5, 6, 7, 8],
- 'possible_implicit_valence_list': [0, 1, 2, 3, 4, 5, 6],
- 'possible_degree_list': [0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10],
- 'possible_bonds': [
- Chem.rdchem.BondType.SINGLE,
- Chem.rdchem.BondType.DOUBLE,
- Chem.rdchem.BondType.TRIPLE,
- Chem.rdchem.BondType.AROMATIC
- ],
- 'possible_bond_dirs': [ # only for double bond stereo information
- Chem.rdchem.BondDir.NONE,
- Chem.rdchem.BondDir.ENDUPRIGHT,
- Chem.rdchem.BondDir.ENDDOWNRIGHT
- ]
- }
-
- def mol_to_graph_data_obj_simple(mol):
- # atoms
- num_atom_features = 2 # atom type, chirality tag
- atom_features_list = []
- for atom in mol.GetAtoms():
- atom_feature = [allowable_features['possible_atomic_num_list'].index(
- atom.GetAtomicNum())] + [allowable_features[
- 'possible_chirality_list'].index(atom.GetChiralTag())]
- atom_features_list.append(atom_feature)
- x = torch.tensor(np.array(atom_features_list), dtype=torch.long)
-
- # bonds
- num_bond_features = 2 # bond type, bond direction
- if len(mol.GetBonds()) > 0: # mol has bonds
- edges_list = []
- edge_features_list = []
- for bond in mol.GetBonds():
- i = bond.GetBeginAtomIdx()
- j = bond.GetEndAtomIdx()
- edge_feature = [allowable_features['possible_bonds'].index(
- bond.GetBondType())] + [allowable_features[
- 'possible_bond_dirs'].index(
- bond.GetBondDir())]
- edges_list.append((i, j))
- edge_features_list.append(edge_feature)
- edges_list.append((j, i))
- edge_features_list.append(edge_feature)
-
- # data.edge_index: Graph connectivity in COO format with shape [2, num_edges]
- edge_index = torch.tensor(np.array(edges_list).T, dtype=torch.long)
-
- # data.edge_attr: Edge feature matrix with shape [num_edges, num_edge_features]
- edge_attr = torch.tensor(np.array(edge_features_list),
- dtype=torch.long)
- else: # mol has no bonds
- edge_index = torch.empty((2, 0), dtype=torch.long)
- edge_attr = torch.empty((0, num_bond_features), dtype=torch.long)
-
- data = Data(x=x, edge_index=edge_index, edge_attr=edge_attr)
- return data

二、模型
预训练时使用到的GIN层和GIN模型,注意这里一定要与预训练的模型一致, context部分我们使用的是GIN模型,直接加载之前的参数即可,这一部分跳过了。
在预训练GIN模型之后要接一个线性层,组成我们的用于分子性质预测整个模型GNN_graphpred。线性层如下:
- #预训练GIN模型与线性层组合成为预测模型
- class GNN_graphpred(torch.nn.Module):
- '''
- 使用预训练相同的结构的gnn,并添加简单的线性层
- '''
- def __init__(self, pre_model, pre_model_files, graph_pred_linear, drop_ratio=0.05, graph_pooling = "mean", if_pretrain=True):
- super(GNN_graphpred, self).__init__()
-
- self.drop_layer = torch.nn.Dropout(p=drop_ratio)
- self.gnn = pre_model
- self.pre_model_files = pre_model_files
-
- #Different kind of graph pooling
- if graph_pooling == "sum":
- self.pool = global_add_pool
- elif graph_pooling == "mean":
- self.pool = global_mean_pool
- elif graph_pooling == "max":
- self.pool = global_max_pool
- elif graph_pooling == "attention":
- if self.JK == "concat":
- self.pool = GlobalAttention(gate_nn = torch.nn.Linear((self.num_layer + 1) * emb_dim, 1))
- else:
- self.pool = GlobalAttention(gate_nn = torch.nn.Linear(emb_dim, 1))
- else:
- raise ValueError("Invalid graph pooling type.")
-
- self.graph_pred_linear = graph_pred_linear #线性层
-
- #加载预训练模型参数:
- if if_pretrain:
- self.from_pretrained()
- self.gnn = self.gnn.eval() # 预训练模型不在参与训练?
-
- def from_pretrained(self,):
- '''
- 加载预训练好的参数
- '''
- #self.gnn = GNN(self.num_layer, self.emb_dim, JK = self.JK, drop_ratio = self.drop_ratio)
- self.gnn = torch.load(self.pre_model_files)
- self.gnn = self.gnn.eval() #预训练模型部分不参与训练
-
- def forward(self, data):
- batch = data.batch
- node_representation = self.gnn(data)
- result = self.pool(node_representation, batch)
- result = self.drop_layer(result)
- result = self.graph_pred_linear(result)
- return result

三、训练过程
单次epoch的训练函数:
使用with torch.no_grad():对测试集进行预测,避免在迭代过程中,显存逐渐增大。
- #单次epcoh训练函数
- def train(model, device, loader_train, loader_test, optimizer, criterion):
- loss_train = []
- r2_train = []
- corr_train = []
- loss_test = []
- r2_test = []
- corr_test = []
-
- model.train()
- for step, batch in enumerate(tqdm(loader_train, desc="Iteration")):
- batch = batch.to(device)
- pred = model(batch)
- y = batch.y.view(pred.shape).to(torch.float64)
- R2 = torch.sum((pred - torch.mean(y))**2) / torch.sum((y - torch.mean(y))**2)
-
- #Whether y is non-null or not.
- is_valid = y**2 > 0
- #Loss matrix
- loss_mat = criterion(pred.double(), (y+1)/2)
- #loss matrix after removing null target
- loss_mat = torch.where(is_valid, loss_mat, torch.zeros(loss_mat.shape).to(loss_mat.device).to(loss_mat.dtype))
-
- optimizer.zero_grad()
- loss = torch.sum(loss_mat)/torch.sum(is_valid) - 0.3 * R2 + 0.3 #添加0.3的R2作为损失
- loss.backward()
- optimizer.step()
-
- #计算预测值与真实值的R2
- pred = pred.detach().cpu().reshape(-1).numpy()
- y = y.detach().cpu().reshape(-1).numpy()
- # r2 = 1 - np.sum((y - pred)**2) / np.sum((y - np.mean(y))**2)
- r2 = np.sum((pred - np.mean(y))**2) / np.sum((y - np.mean(y))**2)
- # r2 = r2_score(y, pred)
- corr = np.corrcoef(y, pred)[0,1]
- loss = loss.detach().cpu().numpy()
- loss_train.append(loss)
- r2_train.append(r2)
- corr_train.append(corr)
- with torch.no_grad():
- for step, batch in enumerate(tqdm(loader_test, desc="Iteration")):
- batch = batch.to(device)
- pred = model(batch)
- y = batch.y.view(pred.shape).to(torch.float64)
- R2 = torch.sum((pred - torch.mean(y))**2) / torch.sum((y - torch.mean(y))**2)
-
- #Whether y is non-null or not.
- is_valid = y**2 > 0
- #Loss matrix
- loss_mat = criterion(pred.double(), (y+1)/2)
- #loss matrix after removing null target
- loss_mat = torch.where(is_valid, loss_mat, torch.zeros(loss_mat.shape).to(loss_mat.device).to(loss_mat.dtype))
- loss = torch.sum(loss_mat)/torch.sum(is_valid) - 0.3 * R2 + 0.3 #添加0.3的R2作为损失
- loss_test_ = loss.detach().cpu().numpy()
- #计算预测值与真实值的R2
- pred = pred.detach().cpu().reshape(-1).numpy()
- y = y.detach().cpu().reshape(-1).numpy()
- r2_test_ = np.sum((pred - np.mean(y))**2) / np.sum((y - np.mean(y))**2)
- # r2_test_ = r2_score(y, pred)
- corr_test_ = np.corrcoef(y, pred)[0,1]
- loss_test.append(loss_test_)
- r2_test.append(r2_test_)
- corr_test.append(corr_test_)
- l = len(loss_train)
- return sum(loss_train)/l, sum(r2_train)/l, sum(corr_train)/l, sum(loss_test)/l, sum(r2_test)/l, sum(corr_test)/l

接下来,就要比较,有预训练和没有预训练的差别,代码如下:
先使用sklearn的train_test_split函数,将监督学习的ESOL等数据集随机划分为训练集和测试集,用于模型性能检测。分别比较预训练和没有预训练的差异。
- if __name__ == '__main__':
- #训练次数
- epoches = 1000
- # 划分数据集,训练集和测试集,要注意PYG的数据存储形式
- data = pd.read_csv('dataset/lipophilicity/raw/Lipophilicity.csv')
- data_train, data_test = train_test_split(data, test_size=0.25, random_state=88)
- data_train.to_csv('dataset/lipophilicity/raw/lipophilicity-train.csv',index=False)
- data_test.to_csv('dataset/lipophilicity/raw/lipophilicity-test.csv',index=False)
- #训练集
- dataset_train = MoleculeDataset(root="dataset/lipophilicity", dataset='lipophilicity-train')
- loader_train = DataLoader(dataset_train, batch_size=64, shuffle=True, num_workers = 8)
- #测试集
- dataset_test = MoleculeDataset(root="dataset/lipophilicity", dataset='lipophilicity-test')
- loader_test = DataLoader(dataset_test, batch_size=64, shuffle=True, num_workers = 8)
-
- '''
- 有预训练条件下
- '''
- #定义使用预训练GIN模型的模型
- pre_model = GNN(7,512) #参数要和预训练的一致,模型结构先实例化一遍
- #线性层
- linear_model = Graph_pred_linear(512, 256, 1)
- #连成新的预测模型
- model = GNN_graphpred(pre_model=pre_model, pre_model_files='Context_Pretrain_Gat.pth', graph_pred_linear=linear_model)
- model = model.to(device)
- #优化器与损失函数
- optimizer = optim.Adam(model.parameters(), lr=0.001, weight_decay=1e-4) # 仅训练model的graph_pred_linear层
- criterion = torch.nn.MSELoss()
- #训练过程
- log_loss = []
- log_r2 = []
- log_corr = []
- log_loss_test = []
- log_r2_test = []
- log_corr_test = []
- for epoch in range(1, epoches):
- print("====epoch " + str(epoch))
- loss, r2, corr, loss_test, r2_test, corr_test = train(model, device, loader_train, loader_test, optimizer, criterion)
- log_loss.append(loss)
- log_r2.append(r2)
- log_corr.append(corr)
- log_loss_test.append(loss_test)
- log_r2_test.append(r2_test)
- log_corr_test.append(corr_test)
- print('loss:{:.4f}, r2:{:.4f}, corr:{:.4f}, loss_test:{:.4f}, r2_test:{:.4f}, corr_test:{:.4f}'.format(loss, r2, corr, loss_test, r2_test, corr_test))
- #保存整个模型
- torch.save(model, "context_pretrian_supervised.pth")
- torch.save(model.state_dict(), "context_pretrian_supervised_para.pth")
- #保存训练过程
- np.save("Context_Supervised_log_train_loss.npy", log_loss)
- np.save("Context_Supervised_log_train_corr.npy", log_corr)
- np.save("Context_Supervised_log_train_r2.npy", log_r2)
- np.save("Context_Supervised_log_train_loss_test.npy", log_loss_test)
- np.save("Context_Supervised_log_train_corr_test.npy", log_corr_test)
- np.save("Context_Supervised_log_train_r2_test.npy", log_r2_test)
- #对测试集的预测
- y_all = []
- y_pred_all = []
- for step, batch in enumerate(loader_test):
- batch = batch.to(device)
- pred = model(batch)
- y = batch.y.view(pred.shape).to(torch.float64)
- pred = list(pred.detach().cpu().reshape(-1).numpy())
- y = list(y.detach().cpu().reshape(-1).numpy())
- y_all = y_all + y
- y_pred_all = y_pred_all + pred
- sns.regplot(y_all, y_pred_all, label='pretrain')
- plt.ylabel('y true')
- plt.xlabel('predicted')
- plt.legend()
- plt.savefig('Context_Supervised_Test_curve.png') #保存图片
- plt.cla()
- plt.clf()
-
- '''
- 没有预训练的条件下
- '''
- pre_model = GNN(7,512) #参数要和预训练的一致
- #线性层
- linear_model = Graph_pred_linear(512, 256, 1)
- #连成新的模型
- model = GNN_graphpred(pre_model=pre_model, pre_model_files='Context_Pretrain_Gat.pth',
- graph_pred_linear=linear_model, if_pretrain=False) # if_pretrain控制不使用预训练的权重
- model = model.to(device)
- optimizer = optim.Adam(model.parameters(), lr=0.001, weight_decay=1e-4)
- criterion = torch.nn.MSELoss()
- un_log_loss = []
- un_log_r2 = []
- un_log_corr = []
- un_log_loss_test = []
- un_log_r2_test = []
- un_log_corr_test = []
-
- for epoch in range(1, epoches):
- print("====epoch " + str(epoch))
- loss, r2, corr, loss_test, r2_test, corr_test = train(model, device, loader_train, loader_test, optimizer, criterion)
- un_log_loss.append(loss)
- un_log_r2.append(r2)
- un_log_corr.append(corr)
- un_log_loss_test.append(loss_test)
- un_log_r2_test.append(r2_test)
- un_log_corr_test.append(corr_test)
- print('loss:{:.4f}, r2:{:.4f}, corr:{:.4f}, loss_test:{:.4f}, r2_test:{:.4f}, corr_test:{:.4f}'.format(loss, r2, corr, loss_test, r2_test, corr_test))
- #对测试集的预测
- y_all = []
- y_pred_all = []
- for step, batch in enumerate(loader_test):
- batch = batch.to(device)
- pred = model(batch)
- y = batch.y.view(pred.shape).to(torch.float64)
- pred = list(pred.detach().cpu().reshape(-1).numpy())
- y = list(y.detach().cpu().reshape(-1).numpy())
- y_all = y_all + y
- y_pred_all = y_pred_all + pred
- sns.regplot(y_all, y_pred_all, label='unpretrain')
- plt.ylabel('y true')
- plt.xlabel('predicted')
- plt.legend()
- plt.savefig('Derectly_Supervised_Test_curve.png') #保存图片
- plt.cla()
- plt.clf()
- '''
- 保存图片,比较有预训练和没有预训练的差距
- '''
- plt.figure(figsize=(15,6))
- plt.subplot(1,3,1)
- plt.plot(log_loss, label='loss')
- plt.plot(log_loss_test, label='loss_test')
- plt.plot(un_log_loss, label='unpretrain_loss')
- plt.plot(un_log_loss_test, label='unpretrain_loss_test')
- plt.xlabel('Epoch')
- plt.ylabel('MSE Loss')
- plt.legend()
- plt.subplot(1,3,2)
- plt.plot(log_corr, label='corr')
- plt.plot(log_corr_test, label='corr_test')
- plt.plot(un_log_corr, label='unpretrain_corr')
- plt.plot(un_log_corr_test, label='unpretrain_corr_test')
- plt.xlabel('Epoch')
- plt.ylabel('Corr')
- plt.legend()
- plt.subplot(1,3,3)
- plt.plot(log_r2[1:], label='r2')
- plt.plot(log_r2_test[1:], label='r2_test')
- plt.plot(un_log_r2[1:], label='unpretrain_r2')
- plt.plot(un_log_r2_test[1:], label='unpretrain_r2_test')
- plt.ylim(0,1)
- plt.xlabel('Epoch')
- plt.ylabel('R2')
- plt.legend()
- plt.savefig('Comversion_Train_process.png')

结果文件中,
Comversion_Train_process.png:损失函数、相关系数、R2的对比;
Derectly_Supervised_Test_curve.png:不经过预训练,直接从头训练的最后拟合曲线;
Context_Supervised_Test_curve.png:预训练模型,最后的拟合曲线;
*.pth:模型。
四、结果
使用context预训练的GIN模型esol数据集,100 epochs:
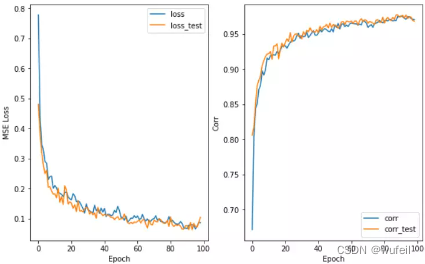
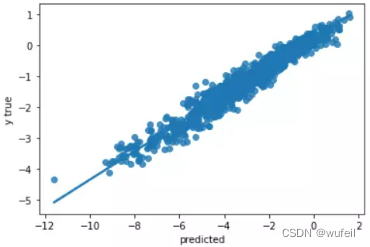
没有使用预训练的GIN模型esol数据集:
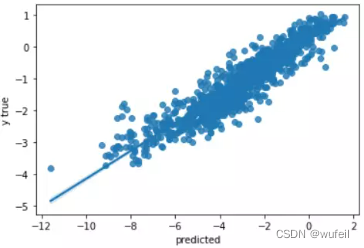
从结果来看,不管是从训练集还是测试集的loss或者相关系数来看,context预训练的结果很明显。在下图500个循环中,也很明显。
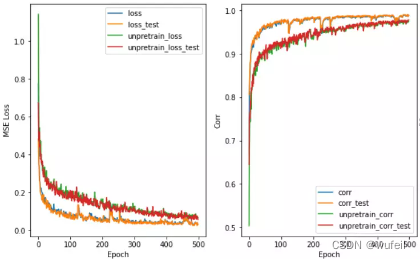
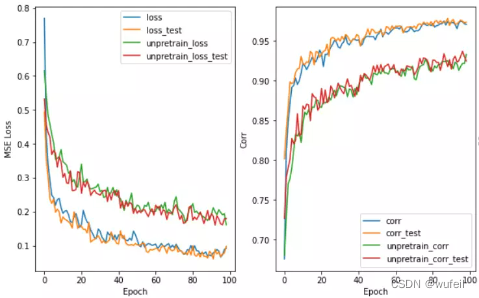
在Lipophilicity数据集上效果也是很明显,如下图:
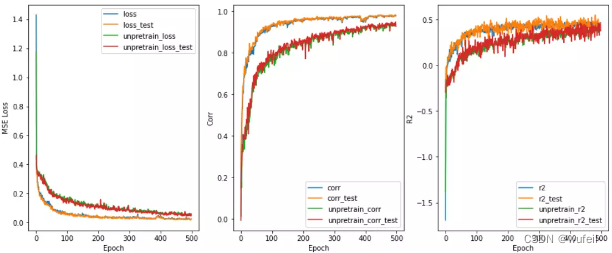
在下图1000个循环中,也很明显。说明经过预训练可以减少训练的迭代次数,减少过拟合。
下图中左为预训练模型的测试集拟合曲线,右图为未预训练模型的测试集拟合曲线。可以看出,经过预训练以后,模型性能确实得到了较大的提高。
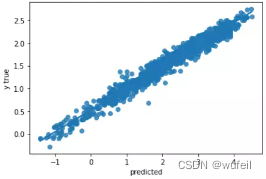
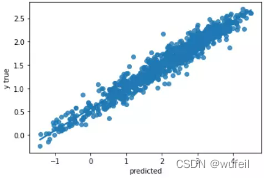
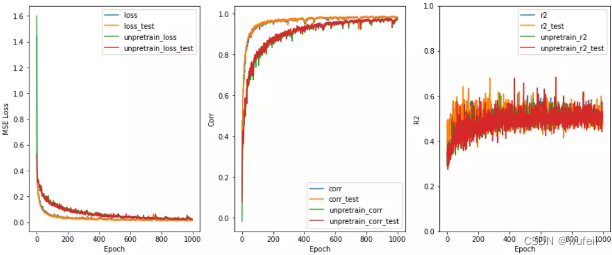
目前存在的问题是:相关系数已经很高了,高达0.99,但是R2却只有0.5左右。所以,我们考虑将损失函数中,添加少量的R2,添加比例为0.3,这里建议不超过0.5,否则一开始的R2就会接接近1,在运行过程中,R2和corr的波动也会很大。结果如下:
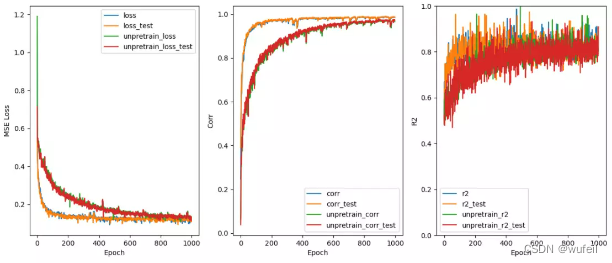
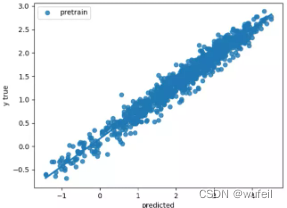
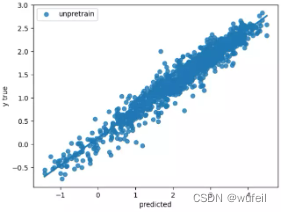
从上图结果来看,预训练还是有效果的。如果迭代次数很多,效果不会非常明显,模型性能提升有限。
五、源代码下载
链接:https://pan.baidu.com/s/14cxHjU2zwzkqPfwwfuSx0Q
提取码:795y


