- 1黑猴子的家:Redis 持久化 之 RDB_dump.rdb.bak
- 2重要一个设置 adb connect 由于目标计算机积极拒绝,无法连接。 (10061)_雷电模拟器 adb 连接 由于目标计算机积极拒绝,无法连接。 (10061)
- 3【ICML 2020】REALM: Retrieval-Augmented Language Model PreTraining
- 4「最新版」SpringBoot3.2.0官方教程实践01-构建一个RESTful Web服务_spring boot 3.2.0
- 5区块链和人工智能的关系以及经典案例_区块链和ai的关系
- 6通信算法之四十六:OFDM系统波形设计_shi算法 ofdm
- 7最新最详细的Python开发环境搭建以及PyCharm的安装配置教程【图+文】_pycharm最后的rebootnow
- 8Leetcode- 使字符串平衡的最小交换次数
- 9Vivado 3-8译码器 4-16译码器_4输入十六进制显示译码器vivado
- 10爬虫练习:Selenium使用案例
GWAS分析中SNP解释百分比PVE | 第二篇,GLM模型中如何计算PVE?
赞
踩
上一篇,介绍了一下显著性的SNP,他们的解释表型变异百分比(PVE)之和,为何可能大于1.
https://yijiaobani.blog.csdn.net/article/details/122093602
这篇我们介绍一下GLM模型中,PVE的计算方法。
1. 数据描述
协变量是PCA的前三个,数据具体如下:
表型数据:
1641 1641 153.973423
1642 1642 113.119301
1643 1643 77.094801
1644 1644 89.293866
1645 1645 94.511433
1646 1646 134.410909
1647 1647 121.246759
1648 1648 45.699443
1649 1649 67.786229
1650 1650 102.225648
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
协变量数据
1641 1641 0.0633225 0.0285328 -0.00734127
1642 1642 0.0684039 0.0212648 -0.000664363
1643 1643 0.0609595 0.0242615 -0.00206211
1644 1644 0.0636631 0.0241012 -0.00275062
1645 1645 0.0741456 0.0293644 0.00068775
1646 1646 0.114417 0.0443451 -0.0408687
1647 1647 0.111599 0.0400401 -0.0320522
1648 1648 0.100862 0.0344925 -0.0386237
1649 1649 0.107986 0.028411 -0.0312307
1650 1650 0.108836 0.0377951 -0.0314308
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
基因型数据:
FID IID PAT MAT SEX PHENOTYPE M4_A M6_T M9_T M11_A M17_A M19_A M22_A M24_A M27_A M31_ 1641 1641 0 0 0 -9 0 1 2 1 0 0 1 0 0 0 0 1 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 1 1 1642 1642 0 0 0 -9 1 1 1 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 1643 1643 0 0 0 -9 1 0 1 1 0 0 1 1 1 1 0 0 0 0 0 0 1 1 0 1 0 0 0 0 0 0 0 0 0 1 1 1 1 1644 1644 0 0 0 -9 1 0 1 1 0 0 1 1 1 1 0 0 0 0 0 0 1 1 0 1 0 0 0 0 0 0 0 0 0 1 1 1 1 1645 1645 0 0 0 -9 0 1 2 1 0 0 1 0 0 0 0 1 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 1 1 1646 1646 0 0 0 -9 1 0 0 0 0 0 0 2 2 2 1 0 0 0 0 0 1 1 0 1 0 0 0 0 0 0 0 0 0 1 1 0 0 1647 1647 0 0 0 -9 0 1 1 0 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0 0 1648 1648 0 0 0 -9 1 0 0 0 0 0 0 1 1 2 1 0 0 0 0 0 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 1649 1649 0 0 0 -9 0 1 1 0 0 0 0 1 1 1 1 1 1 0 0 0 0 0 0 1 0 0 0 0 0 0 0 0 0 1 0 0 0 1650 1650 0 0 0 -9 1 0 0 0 0 0 0 1 1 2 1 0 0 0 0 0 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 1651 1651 0 0 0 -9 1 0 0 0 0 0 0 1 1 1 0 0 0 0 0 0 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 1652 1652 0 0 0 -9 0 1 1 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0 0 1653 1653 0 0 0 -9 1 0 0 0 0 0 0 1 1 1 0 0 0 0 0 0 1 1 0 0 0 0 0 0 1 1 1 1 1 1 1 0 0 1654 1654 0 0 0 -9 0 1 1 0 0 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 1 1 1 1 1 1 0 0 0 1655 1655 0 0 0 -9 2 0 0 0 0 0 1 1 1 1 0 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 0 0 1 0 0 1656 1656 0 0 0 -9 2 0 0 0 1 1 2 2 2 2 2 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1657 1657 0 0 0 -9 2 0 0 0 1 1 2 2 2 2 2 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1658 1658 0 0 0 -9 2 0 0 0 1 1 2 2 2 2 2 0 1 1 1 1 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
2. GAPIT软件分析GLM模型
GAPIT软件安装,见:如何安装GAPIT软件:https://zhuanlan.zhihu.com/p/268327005
数据及代码下载,请关注公众号:育种数据分析之放飞自我,进入知识星球进行相关下载和学习
分析代码如下:
library(data.table) raw = fread("plink.raw",header=T) raw[1:10,1:10] map = fread("recode.map",header=F) head(map) rr = raw[,!c(1,3:6)] names(rr) = c("taxa",map$V2) mm = map[,c(2,1,4)] names(mm) = c("Name","Chromosome","Position") head(mm) rr[1:10,1:10] fwrite(rr,"mdp_numeric.txt",sep="\t") fwrite(mm,"mdp_SNP_information.txt",sep="\t") # # GWAS 分析 library(data.table) source("http://zzlab.net/GAPIT/GAPIT.library.R") source("http://zzlab.net/GAPIT/gapit_functions.txt") myGd = fread("mdp_numeric.txt",header=T,data.table = F) myGM = fread("mdp_SNP_information.txt",header = T,data.table=F) myY = fread("dat_plink.txt",data.table = F) head(myY) covar = fread("cov_plink.txt",data.table = F)[,-1] names(covar)[1] = "Taxa" head(covar) myGAPIT = GAPIT(Y = myY[,c(1,3)],GD = myGd, GM = myGM, # PCA.total=3, CV = covar, model="GLM")
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
- 24
- 25
- 26
- 27
- 28
- 29
- 30
- 31
- 32
- 33
- 34
- 35
- 36
- 37
- 38
- 39
- 40
- 41
注意:
- 这里使用的是plink格式的表型和PCA结果,使用的是plink.raw文件为基因型数据
- 将其转化为gapit软件需要的格式
- 定义基因型和位置信息,定义表型,定义协变量,定义模型为GLM
- 结果文件为:
GAPIT.GLM.V3.GWAS.Results.csv
3. GLM模型结果文件
结果文件如下:
包括:
- SNP名称
- 染色体
- 物理位置
- P值 # 重点关注
- maf
- Rsquare.of.Model.without.SNP # 重点关注
- Rsquare.of.Model.with.SNP # 重点关注
SNP,Chromosome,Position ,P.value,maf,nobs,Rsquare.of.Model.without.SNP,Rsquare.of.Model.with.SNP,FDR_Adjusted_P-values,effect
M57157,12,44,0.0000000265778189052299,0.375,1000,0.0178325320026741,0.0488405467802565,0.000799726570858368,7.5184135943861
M57155,12,44,0.000000149990719134015,0.3275,1000,0.0178325320026741,0.0454335956044116,0.00115674059672426,7.24232622801506
M57156,12,44,0.000000149990719134015,0.3275,1000,0.0178325320026741,0.0454335956044116,0.00115674059672426,7.24232622801506
M98663,20,73,0.00000015377076726145,0.222,1000,0.0178325320026741,0.0453847644903734,0.00115674059672426,-7.81213756957444
M73233,15,64,0.000000512284630336812,0.2255,1000,0.0178325320026741,0.0430299031398552,0.00290006393781271,-7.52596405927775
M8452,2,69,0.000000578277953701437,0.435,1000,0.0178325320026741,0.0427934752525868,0.00290006393781271,-6.51962588787404
M45998,10,20,0.000000768626039613293,0.411,1000,0.0178325320026741,0.0422387923854602,0.00307979999092427,-6.69120649416338
M82957,17,57,0.00000112940850557263,0.1685,1000,0.0178325320026741,0.0414897712489706,0.00307979999092427,8.48568078650835
M82958,17,57,0.00000112940850557263,0.1685,1000,0.0178325320026741,0.0414897712489706,0.00307979999092427,8.48568078650835
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
这里,
- P值 为SNP的P值,我们根据它判断显著性,并根据它进行QQ图和曼哈顿图的绘制
- Rsquare.of.Model.without.SNP # 这个是单位点不包括次SNP的解释百分比(R方)
- Rsquare.of.Model.with.SNP # 这个是单位点包括此SNP的解释百分比(R方)
上面两者之差,即为该SNP的解释百分比(PVE)
$$SNP的PVE = Rsquare.of.Model.with.SNP - Rsquare.of.Model.without.SNP$$
- 1
4. 我们将结果读入R语言计算
首先读入R语言:
r$> re = fread("GAPIT.GLM.V3.GWAS.Results.csv")
r$> head(re)
SNP Chromosome Position P.value maf nobs Rsquare.of.Model.without.SNP Rsquare.of.Model.with.SNP FDR_Adjusted_P-values effect
1: M57157 12 44 2.657782e-08 0.3750 1000 0.01783253 0.04884055 0.0007997266 7.518414
2: M57155 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.242326
3: M57156 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.242326
4: M98663 20 73 1.537708e-07 0.2220 1000 0.01783253 0.04538476 0.0011567406 -7.812138
5: M73233 15 64 5.122846e-07 0.2255 1000 0.01783253 0.04302990 0.0029000639 -7.525964
6: M8452 2 69 5.782780e-07 0.4350 1000 0.01783253 0.04279348 0.0029000639 -6.519626
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
然后我们根据上面的公式,手动计算PVE值:
r$> re$PVE = re$Rsquare.of.Model.with.SNP - re$Rsquare.of.Model.without.SNP
r$> head(re)
SNP Chromosome Position P.value maf nobs Rsquare.of.Model.without.SNP Rsquare.of.Model.with.SNP FDR_Adjusted_P-values effect PVE
1: M57157 12 44 2.657782e-08 0.3750 1000 0.01783253 0.04884055 0.0007997266 7.518414 0.03100801
2: M57155 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.242326 0.02760106
3: M57156 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.242326 0.02760106
4: M98663 20 73 1.537708e-07 0.2220 1000 0.01783253 0.04538476 0.0011567406 -7.812138 0.02755223
5: M73233 15 64 5.122846e-07 0.2255 1000 0.01783253 0.04302990 0.0029000639 -7.525964 0.02519737
6: M8452 2 69 5.782780e-07 0.4350 1000 0.01783253 0.04279348 0.0029000639 -6.519626 0.02496094
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
根据PVE的大小,进行排序,降序:
r$> re %>% arrange(-PVE)
SNP Chromosome Position P.value maf nobs Rsquare.of.Model.without.SNP Rsquare.of.Model.with.SNP FDR_Adjusted_P-values effect PVE
1: M57157 12 44 2.657782e-08 0.3750 1000 0.01783253 0.04884055 0.0007997266 7.5184135944 3.100801e-02
2: M57155 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.2423262280 2.760106e-02
3: M57156 12 44 1.499907e-07 0.3275 1000 0.01783253 0.04543360 0.0011567406 7.2423262280 2.760106e-02
4: M98663 20 73 1.537708e-07 0.2220 1000 0.01783253 0.04538476 0.0011567406 -7.8121375696 2.755223e-02
5: M73233 15 64 5.122846e-07 0.2255 1000 0.01783253 0.04302990 0.0029000639 -7.5259640593 2.519737e-02
---
30086: M58296 12 66 9.996606e-01 0.2700 1000 0.01783253 0.01783253 0.9997934933 0.0006040638 1.785355e-10
30087: M751 1 15 9.998043e-01 0.2435 1000 0.01783253 0.01783253 0.9998960441 0.0003736382 5.935820e-11
30088: M21736 5 35 9.998296e-01 0.3020 1000 0.01783253 0.01783253 0.9998960441 0.0002955665 4.500680e-11
30089: M80677 17 12 9.999131e-01 0.3830 1000 0.01783253 0.01783253 0.9999300138 -0.0001448323 1.170910e-11
30090: M57593 12 52 9.999300e-01 0.2735 1000 0.01783253 0.01783253 0.9999300138 -0.0001300807 7.589301e-12
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
可以看到,结果就给出了PVE从大到小的排序结果。
这里,我们注意到,前三个SNP的PVE分别是:
- 3.1%
- 2.76%
- 2.76%
而且,这三个SNP都在12号染色体的44个位置,因为这三个位点离得很近,都达到显著水平,PVE也很大,那我们可以认为附件有一个基因,显著影响表型。
这里,我们对PVE进行求和:
r$> sum(re$PVE)
[1] 57.6114
- 1
- 2
- 3
可以看到,总的PVE之和为57.611,远远大于1,是因为有些位点高度LD,所以PVE之和就很大。相关问题在 GWAS分析中SNP解释百分比PVE | 第一篇,SNP解释百分比之和为何大于1?中有过介绍。
5. 用R语言如何计算?
简单来说,就是单位点的回归分析,计算R方。
这里,我们用同样的数据,在R中进行GLM的GWAS分析。
代码如下:
library(data.table) geno = fread("plink.raw")[,!c(1,3:6)] map = fread("recode.map") m012 = geno names(m012) = c("IID",map$V2) phe = fread("phe.txt") cov1= fread("tcov.txt") dat = left_join(phe,cov1,by = c("V1"="V1")) %>% dplyr::select(IID =1, y =2, pc1=3,pc2=4,pc3=5) dat1 = left_join(dat,m012,by="IID") # dat1$M57157 = as.factor(dat1$M57157) mod_1 = lm(y ~ 1+pc1 + pc2 + pc3 + M57157,data=dat1);summary(mod_1) summary(mod_1)$r.squared mod_2 = lm(y ~ 1+pc1 + pc2 + pc3 ,data=dat1);summary(mod_2) summary(mod_2)$r.squared
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
这里,我们将PCA的前三个作为协变量加到回归分析汇总。
包含SNP的回归分析:
r$> mod_1 = lm(y ~ 1+pc1 + pc2 + pc3 + M57157,data=dat1);summary(mod_1) Call: lm(formula = y ~ 1 + pc1 + pc2 + pc3 + M57157, data = dat1) Residuals: Min 1Q Median 3Q Max -85.094 -19.174 -0.443 18.218 79.181 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 96.408 1.320 73.048 < 2e-16 *** pc1 -108.543 27.603 -3.932 9.00e-05 *** pc2 28.740 28.981 0.992 0.3216 pc3 58.098 27.890 2.083 0.0375 * M57157 7.518 1.320 5.695 1.62e-08 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 27.6 on 995 degrees of freedom Multiple R-squared: 0.04884, Adjusted R-squared: 0.04502 F-statistic: 12.77 on 4 and 995 DF, p-value: 3.838e-10
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
- 23
可以看到,R方为0.04884,也可以这样提取:
r$> summary(mod_1)$r.squared
[1] 0.04884055
- 1
- 2
- 3
不包含SNP的回归分析:
r$> mod_2 = lm(y ~ 1+pc1 + pc2 + pc3 ,data=dat1);summary(mod_2) Call: lm(formula = y ~ 1 + pc1 + pc2 + pc3, data = dat1) Residuals: Min 1Q Median 3Q Max -89.495 -19.318 -0.099 18.649 85.448 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 102.0467 0.8864 115.129 < 2e-16 *** pc1 -111.8205 28.0295 -3.989 7.11e-05 *** pc2 -21.6604 28.0295 -0.773 0.44 pc3 35.1350 28.0295 1.254 0.21 --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 28.03 on 996 degrees of freedom Multiple R-squared: 0.01783, Adjusted R-squared: 0.01487 F-statistic: 6.028 on 3 and 996 DF, p-value: 0.0004546
- 1
- 2
- 3
- 4
- 5
- 6
- 7
- 8
- 9
- 10
- 11
- 12
- 13
- 14
- 15
- 16
- 17
- 18
- 19
- 20
- 21
- 22
这里的R方为:0.01783
也可以这样提取:
r$> x2 = summary(mod_2)$r.squared
summary(mod_2)$r.squared
[1] 0.01783253
- 1
- 2
- 3
- 4
那么SNP:M57157的PVE为:3.1%
r$> x1 - x2
[1] 0.03100801
- 1
- 2
- 3
对比我们GAPIT的PVE结果:
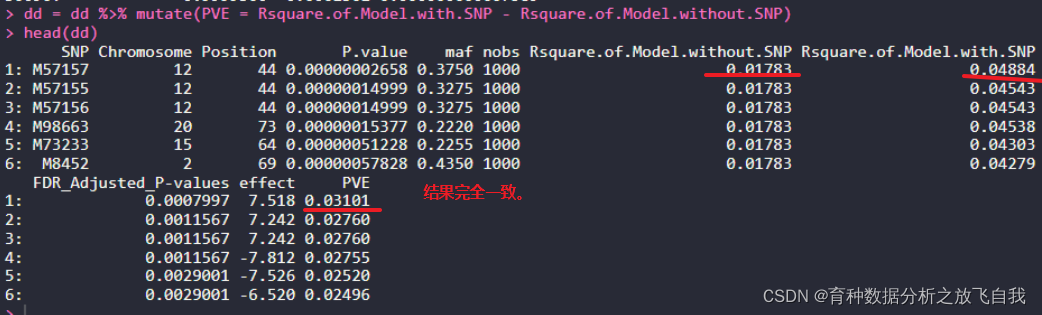
结果完全一致。
这里,一般线性模型中,可以针对显著性的SNP,进行单位点回归分析,计算PVE。对于混合线性模型,也可以将显著性位点提取,进行R语言的手动计算,这个也是PVE计算的一种方法。
混合线性模型中,还有其它的计算方法,我们后面进行介绍,欢迎继续关注我。
6. 数据和代码下载方法
数据及代码下载,请关注公众号:育种数据分析之放飞自我,进入知识星球进行相关下载和学习




